Introduction
This article shows how enzymatic activity can be effectively determined by means of the Eppendorf BioSpectrometer. The preliminary measurements were carried out by utilizing the “single λ-continuous” method for optimized measurement process. Based on these outcomes, the exact measurements of the enzyme activity were carried out. Linear regression was used to measure enzyme activities.
A coupled reaction of a glucose-6-phosphate-dehydrogenase from the baker’s yeast and a hexokinase was utilized for the measurements. It is also possible to demonstrate the temperature dependence of the coupled reaction, as the Eppendorf BioSpectrometer kinetic is provided with a cuvette chamber that can be controlled by temperature. Maximum activity was observed when the temperature was 37°C.
Metabolic Significance of the Hexokinase Reaction
Glycolysis
Glycolysis is a cellular process in which glucose is enzymatically degraded in the cytoplasm. This process occurs in almost all living beings. Two molecules of pyruvate originate from glucose, in a stepwise manner.
One of the glucose molecules provides the equivalents of 2 ATP and 2 redox in the form of NADPH2. Next, pyruvate enters the citrate cycle, which is an additional elementary metabolic pathway, so as to provide onward reduction equivalents for the respiratory chain. These reduction equivalents are then given to oxygen. H2O and CO2 are the final degradation products.
Activation of Glucose via the Hexokinase Reaction
The degradation of glucose supports the respiration cycle of all living organisms. Glucose has to be activated i.e. has to be made suitable for degradation, before the degradation process begins. This action involves phosphorylation by enzyme hexokinase, forming glucose-6-phosphate with the expenditure of ATP. The glucose-6-phosphate is the initial substrate of glycolysis (Figure 1). Additionally, it is also the initial substrate of the pentose phosphate pathway, a key metabolic pathway. This pathway serves to supply redox equivalents including NADPH2, and also many different carbohydrates believed for subsequent pathways of catabolic and biosynthesis.
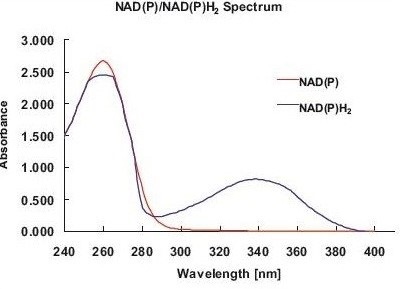
Figure 1. Absorbance spectrum of NADP/ NADPH2. Both components can be easily distinguished by the absorbance maximum of NADPH2 at 340nm. Thus, the activities of NADP/ NADPH2-dependent enzymes are determined via a change in absorbance at 340nm.
Detection of Enzymatic Degradation of Metabolic Enzymes
These metabolic pathways and the enzymes are the subject of biochemical research and education because of their critical significance. Since these comprise the specific hallmark of an enzyme, one of the key aspects is to know the enzymatic activities. A spectrometer, using the principle of photometry, can measure the respective enzymatic activities.
These activities are mainly known via the increase or decrease of NADPH2, as NADPH2 (NADH2) and NADP (NAD) are involved in almost all enzymatic reactions within the central metabolism. NADPH2 and NADP exhibit maximum absorbance at 260nm, although NADPH2 displays another peak at 340nm (Figure 1), allowing for photometric distinction between NADP and NADPH2.
Besides enzyme characterization, enzymatic activity is helpful in determining specific substrates or concentrations of substrates. The hexokinase reaction, mentioned earlier, is useful in determining the concentrations of ATP or glucose. The detection is performed in an indirect manner through the formation of NADPH2 in a coupled reaction using the glucose-6-phosphate-dehydrogenase. The quantity of ATP formed depends on the glucose amount (Figure 2).
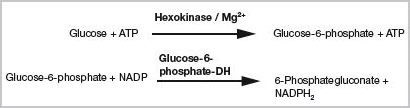
Figure 2. Indirect enzymatic detection of glucose or ATP. Both substances may be determined via a coupled reaction of hexokinase and glucose-6-phosphate-dehydrogenase (coupled test system [1]). The respective amount is calculated from the amount of NADPH2 generated.
The detection reaction shows two uses of the measurements of enzymatic kinetics by utilizing the Eppendorf BioSpectrometer kinetic:
Enzymatic determination of ATP concentration
In this application, the concentration of the substrate is determined through two-point calibration by utilizing the defined amounts of ATP at 37°C. In this process, two measurements are obtained inside the linear range of substrate conversion. In preliminary measurements, the linear range of conversion has to be established.
Determination of enzyme activities
This application helps to test the temperature dependence of the hexokinase reaction (Figure 2). The Eppendorf BioSpectrometer kinetic enables the regulation of sample temperature in a cuvette chamber: a temperature range between 20°C and 42°C can be selected. Since incubation temperature is a factor for enzyme activity, regulation of temperature is vital for accurate activity determination. Thus, the activity of hexokinase enzyme was determined at three different temperatures: 22, 30 and 37°C. Linear regression of the curve is used for exact determination.
Materials
The following materials were used for analysis:
- BioSpectrometer kinetic, IsoPack with IsoRack (Eppendorf)
- ATP (Roche Applied Science, 10127523001)
- Ultra-Mikro cuvette 105.202-QS (Hellma, 105-202-85-40)
- Hexokinase from Saccharomyces cerevisiae (Roche Applied Science, 11426362001)
- NADP (Sigma-Aldrich, 93205-50MG)
- Glucose-6-phosphate-dehydrogenase from Saccharomyces cerevisiae (Roche Applied Science, 10127671001)
- MgCl2 (Sigma-Aldrich, 63069-100ML)
- α-D-Glucose (Sigma-Aldrich,158968-25G)
- Tris buffer pH 7.0 (AppliChem, A5247, 0500)
- Water for molecular biology (AppliChem, A7398,1000)
Methods
Hexokinase Activity Measurements
All hexokinase activity measurements were carried out using the Eppendorf BioSpectrometer. The solutions prepared for the enzyme test are:
- 10mL 0.1M Tris buffer pH 7.0
- 1mL 5mM Glucose
- 1mL 0.1M MgCl2
- 1mL 1mM ATP
- 1mL Hexokinase
- 0.5mL 10mM NADP+
- 2mL Glucose-6-phosphate-dehydrogenase
These solutions were made by utilizing molecular biology-grade water, and were maintained at 4°C in the Eppendorf IsoPack, while the Tris buffer was maintained at ambient temperature. After their usage, the enzymes were immediately moved to the refrigerator. Table 1 shows the components utilized for a single measuring reaction:
Table 1. Components needed for one measuring reaction
|
5μL
|
MgCl2
|
|
5μL
|
NADP
|
|
5μL
|
Glucose
|
|
12.5μL
|
ATP
|
|
121.5μL
|
Tris buffer
|
|
0.5μL
|
Glucose-6-phosphate-dehydrogenase
|
|
Σ = 149.5μL
|
|
An ultra-micro cuvette is shifted into the cuvette chamber, after the solutions are pipetted into the cuvette. For adapting to the reaction temperature, a pre-incubation period for the reactions lasting at least 6 minutes is included as part of the measuring parameters. After starting the measurements, absorbance changes at 340nm were recorded.
After approximately 1 minute, the enzymatic reaction was started by adding 0.5μL hexokinase solution, when further absorbance changes are not detected. The hexokinase and the glucose-6-phosphate-dehydrogenase are added and thoroughly mixed. Measurements were carried out at three different temperatures: 22, 30 and 37°C.
Substrate Determination Via The Hexokinase Test
The components that were used for the hexokinase activity test are also used for substrate determination. 1mM and 0.1mM ATP solutions are used as standards for determining the unknown ATP concentration in a sample.
Settings on the BioSpectrometer Kinetic
Three methods for activity measurements (Figure 3) can be presented by the BioSpectrometer kinetic:
(a) “Single λ-continuous (Single λ-cont)”: This involves, at a defined wavelength, at defined time intervals, and over a defined period of time, simple measurement of absorbance changes; the temperature can be controlled. This method is helpful for preliminary experiments, i.e. determination of the kinetics (speed, linearity range) of a reaction.
(b) “Simple kinetics”: as in (a); in addition, conversion factors and units needed for direct conversion of the absorbance values, can be defined. Two-point measurement, endpoint measurement, or linear regression is available. “Delay” allows programming a delayed measurement action, for e.g. ensuring adequate adaptation of the reaction mixture to the real measurement temperature.
(c) “Advanced kinetics”: as in (b), with an extra opportunity of programming a “reagent blank” and a measurement including a standard. “Reagent Blank” enables verifying the absence of obvious changes in absorbance before the initiation of the reaction. A preliminary reaction was carried out using the “Single λ-cont.” method, for testing the progress of the hexokinase reaction. Selection of parameters was done such that absorbance changes in the reaction mix can be monitored over a prolonged period (10 minutes).
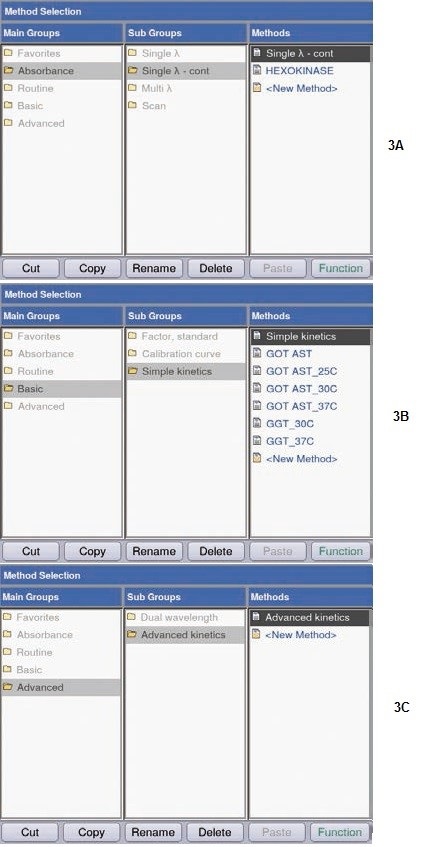
Figure 3. Activity measurement options. (A) Single λ- cont.; (B) Simple kinetics; (C) Advanced kinetics.
The data were collected for every 10 seconds, as reaction speed can undergo rapid changes during enzymatic reactions (Figure 4).
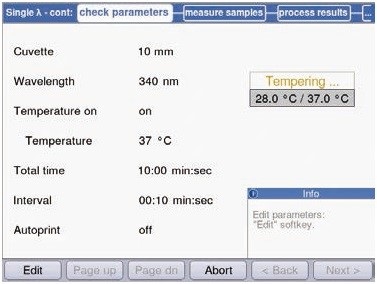
Figure 4. Measurement parameters of preliminary experiments for hexokinase measurements within the method “Single l-cont”.
Based on the preliminary experimental results, the parameters for activity measurements (Figure 5A) as well as substrate determination (Figures 5B and 5C) were fixed. Figure 5 shows the various methods that were opted for substrate concentration vs. activity determination. Linear regression was used to determine hexokinase activity. A fresh measurement was performed every 5 seconds, and total measurement lasted for 6 minutes.
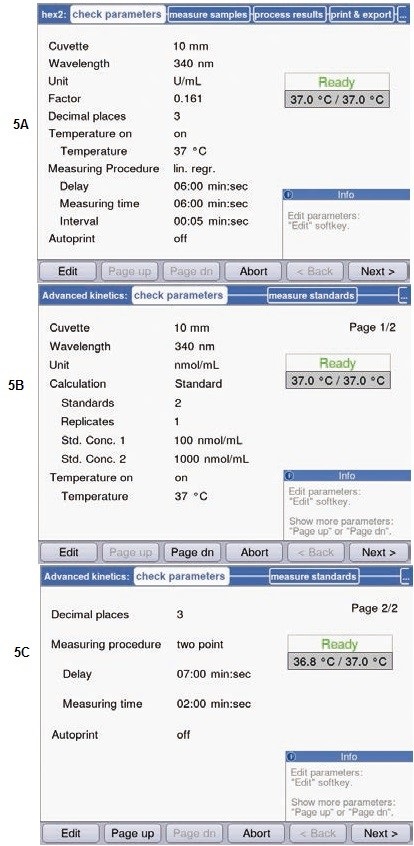
Figure 5. Parameters for determination of activity and substrate concentration. (A) Parameters for determination of hexokinase via linear regression in the method “simple kinetics”. (B) + (C) Parameters for determination of substrate concentration in the method “advanced kinetics” via two point calibration.
The reaction began with the addition of hexokinase, after a warm-up period of 6 minutes. The preliminary experiments were performed by using the “single λ-cont”, and which was the basis for selecting experimental parameters. The formulae for calculating enzyme activity or substrate concentration are given below.
Figure 5A shows the factor which is measured from the molar absorption coefficient for NADPH (6.32 * 103L/(mol * cm)). The Lambert-Beer law postulates:
A= ε*c*l or c=A/ ε*l
Where c= concentration, A= absorption, ε = molar absorption coefficient, l=optical pathway (path length of the cuvette).
For a cuvette having 1cm path length, the formulae to be used are:
c= A/ ε or c= A * 1/ ε or c=A * F, respectively. Therefore, the factor F, having the unit of mol/L or μmol/μL, is the reciprocal value of the molar extinction coefficient (for NADPH2 1.61 * 10-4).
The factor has to be adapted from mol/L for the unit μmol/L, i.e. it has to be multiplied by 1 * 106, for measuring the enzymatic activity U/L or μmol/min * L, respectively, from the absorption measurement. Thus, the factor is 161. Figure 5A shows the conversion factor for the enzymatic activity for 1mL; in this case, the factor is 0.161. The slope is calculated, from which the enzymatic activity is directly derived and displayed subsequent to the measurement.
Determination of Substrate Concentration
For knowing substrate concentration, a measurement is carried out after a pre-incubation period of 7 minutes.
Hexokinase is added, and the reaction mixture was incubated for 2 minutes. As per two-point measurement technique, two measurements are carried out, one at the beginning of the incubation, and the other at the end of it. In order to determine the concentration of sample, two preliminary standard measurements were carried out with substrate of defined concentrations, and the unknown sample concentration is determined from the standards.
Implementation of a Linear Regression on the Biospectrometer Kinetic for the Purpose of Activity Measurements
Since inhibitory factors such as inhibition from the final product or declining concentration of substrate can only be excluded within the range of linearity, the activities that are analyzed are only those which are within the curve’s linear range. For activity measurements that need to be studied utilizing linear regression, the Eppendorf BioSpectrometer kinetic can retroactively adapt the regression curve to the linear curve. For this purpose, after the measurement process, the function “linear regression” in the area “process results” is activated. Thus, the regression curve’s start and end points can be reset (Figure 6).
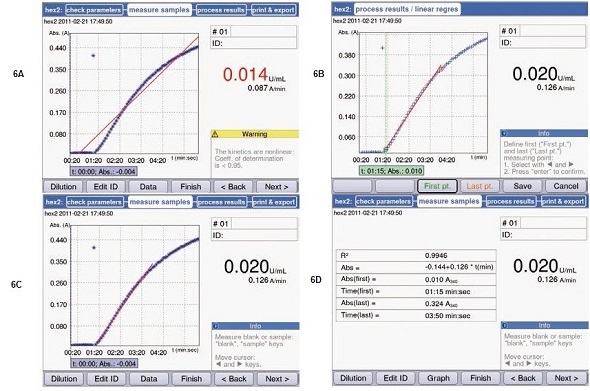
Figure 6. Linear regression analysis following the completion of measurements. (A) Result of a kinetic measurement with linear regression. (B) Start and end point of the regression curve may be reset using the soft keys “First pt” and “Last pt”. (C) New measurement result by way of adjusted regression curve. The result is now displayed in black. (D) All important information pertaining to the regression curve: coefficient of determination, start and end point of the result, regression curve formula.
Results
Preliminary Experiments for Activity Measurements – “Single λ-cont”
When a spectrometer is utilized to measure enzymatic activity, prediction of the course of a reaction is usually not possible. This happens in cases when the measurements were not performed before, or an enzyme’s activity test has to be framed de novo.
In the second case, the enzyme, the necessary substrate, or, if needed, the additive concentrations or co-factors are not known. The speed of the enzymatic reaction, which depends on the components used, is also unknown.
As stated earlier, for a preliminary experiment, the Eppendorf BioSpectrometer kinetic can use the “Single λ-cont” method to test the course of the reaction. Frequent measurements, even after every 5 seconds, can be carried out for a maximum time period of 1 hour.
Data were collected for every 10 seconds for a time period of 10 minutes during the preliminary measurements of hexokinase activity. The sample was under pre-incubation for a minimum period of 6 minutes, before the actual measurement, which enables the sample to adapt itself to the reaction temperature. The reaction began by the addition of hexokinase after 1 minutes.
The aim of the reaction was two-fold: determining the linear range of the curve, and determining the time at which any further major conversion of substrate does not occur. The second objective is vital for substrate determination, because measurement was carried out based on the endpoint approach. Preliminary measurements were then carried out at a temperature of 37°C. Figure 7 shows the results of the measurements.
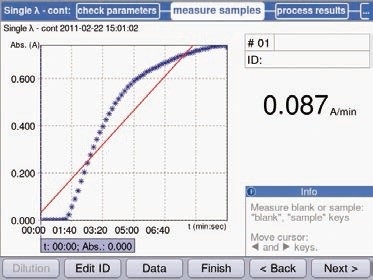
Figure 7. Preliminary measurements of hexokinase using the method “Single λ-cont”. The increase in extinction occurs immediately following addition of the enzyme. The course of the curve is linear for approximately 2 minutes, followed by a flattening of the curve. After 6 minutes, no further significant increase in absorption can be detected.
The parameters for the next experiment were set based on these results. Measurements were limited to 6 minutes, since almost all substrate were converted during this time period. The reaction began after a 1 minute warm-up period. These settings were utilized for the activity tests and also for the determination of substrate concentration.
Determination of an Unknown ATP Concentration – “Advanced Kinetics”
As mentioned, the two-point calibration and the “advanced kinetics” method were to be utilized for finding out the unknown concentration of ATP. For this purpose, two standards were determined at first: a 0.1mM and 1mM ATP solution. Figures 5B and 5C show the measurement parameters. Similar to preliminary experiments, the measurements were carried out at a temperature of 37°C. The results of the standard measurement are shown in Figure 8A. Figure 8B shows that using these standards, the ATP concentration for the unknown sample was found to be 0.79mM.
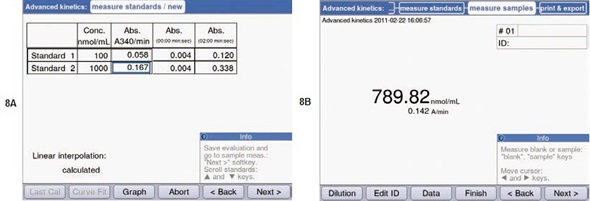
Figure 8. Determination of the ATP concentration using the “Advanced kinetics” methods. (A) Result of the standard measurements. (B) Concentration determined for an unknown sample, calculated from the measured ATP standards.
Determination of Hexokinase Activity at Different Temperatures – “Simple Kinetics”
The BioSpectrometer kinetic has a heatable cuvette chamber, which allows for kinetic measurements to be optimized in relation to the temperature at which they are determined. Subsequent experiments demonstrated the effect of the reaction temperature. For this purpose, hexokinase activity was determined at three temperatures: 22, 30 and 37°C. The measurements were carried out based on the preliminary experiment, i.e. a pre-incubation (“delay”) of 6 minutes, followed by absorption measurements at 340nm at intervals of 5 seconds (also refer to Figure 5A).
Subsequent to a warm-up time of 1 minute, the reaction began with the addition of hexokinase, and was tracked for 5 minutes. Figure 6 shows the synchronization of the regression curve with the linear range of the measured curve, by utilizing linear regression.
Figures 9A-9C show the measurement results at three different temperatures. At 37°C, highest substrate conversion was observed. This activity was over two times of that determined at 22°C. These results are as expected, since the activity maxima of the chosen enzymes should fall between 35°C and 40°C.
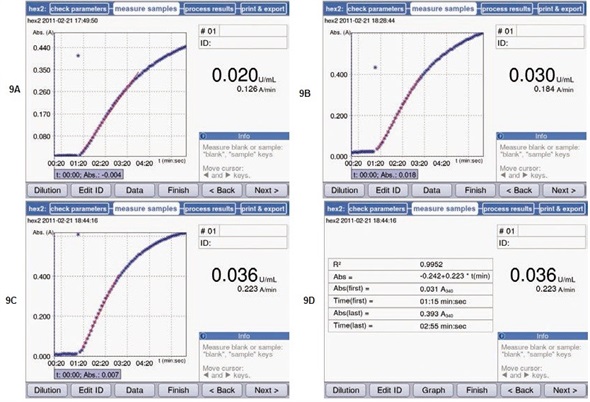
Figure 9. Measurement of hexokinase activity at different temperatures. The results from the measurements were retroactively adapted to the linear range using the regression curve (red line). Further information about the selected range, such as the coefficient of determination of the regression curve, are stored in the area “Data”. (A) Activity at 22°C. (B) Activity at 30°C; (C) Activity at 37°C. (D) Data referring to the selected range, with the example of the measurements at 37°C. Start and end point of the measurement, as well as the respective absorbance values and the calculated coefficient of determination R2.
Conclusion
The Eppendorf BioSpectrometer kinetic is very helpful for the determination of substrate concentrations and for enzyme activity. Using the “Single λ-cont” method, many key parameters including substrate concentrations or optimal enzyme, measurement duration, reaction temperature or linear curve range, can be calculated.
When the course of the reaction is not known or has not been optimized yet, these measurements assume importance. Quick estimation at the completion of substrate conversion is enabled when the molecule concentration is found out by activity measurements. Accurate reaction temperatures can be selected in the heatable cuvette chamber. Accurate finding of the enzyme activity is based on exact temperature selection.
The Eppendorf BioSpectrometer kinetic enables moving the time window retroactively for allowing the measured curve to be adapted to the linear range, and thus enabling its analysis by linear regression. Therefore, enzyme activities can be found out in a reliable manner, since measured curves can be analyzed in a reproducible manner by controlling the coefficient of determination “R2”.
The determination of enzyme activity and substrate concentration is enabled by the choice offered by three measurement methods along with the optional analysis utilizing standard or blank.
Acknowledgements
Produced from materials originally authored by Martin Armbrecht, from Eppendorf AG, Hamburg, Germany.
References
- Friedrich Lottspeich, Joachim W. Engels und Angela Simeon, Bioanalytik, 2nd edition (2008)
- Abrahão-Neto, J., Infanti, P., Vitolo, M., Hexokinase production from S. cerevisiae. Culture conditions. Appl Biochem Biotechnol. 1996 Spring; 57-58:407-12.
About Eppendorf NA
Eppendorf NA, with headquarters in New York, are a subsidiary of Eppendorf AG (Hamburg, Germany) – manufacturer of laboratory instruments and consumables for the life sciences.
To make it easy for labs in the United States to benefit from Eppendorf quality, they provide total customer support and service: taking/tracking orders; product installation and training; application support; calibration services; general product maintenance and repair, and more!
Eppendorf's products
Eppendorf products are used in all types of life science research and testing settings – from basic laboratory applications to highly specialized cell and molecular biology applications. They are highly regarded for their quality design and performance – beginning with extensive research and development, adding state-of-the-art technology and ending with strict quality-controlled manufacturing are what make their products stand out from the rest. It is what has made them a brand you have been able to rely on for over 70 years.
Over the years Eppendorf has improved upon and added to their flagship products – pipettes and pipette tips, centrifuges and microcentrifuge tubes – to now include the most ergonomic liquid handling devices and automated pipetting stations, quiet centrifuges, fast and fully flexible thermal cyclers, sample-protecting deepwell plates, cell manipulation systems and microcapillaries, ultra-low temperature freezers, shakers, incubators and bioprocessing equipment.
Eppendorf's services
Customer Support Representatives are available M–F, 8:30 AM – 8:00 PM (EST).
Local Sales Representatives cover the entire US to give you the fastest best possible attention. They know their products – and better yet, they know your applications. Feel confident in their abilities to assess your needs and determine the best products and systems to give you the best results.
Field Specialists also cover the entire US to provide post-sales installation and training on their specialty systems and automation.
The Applications Hotline is manned by degreed scientists working in their own fully functional life science laboratory. They are on call and email-accessible to help you troubleshoot your application or answer any technical question you may have. The Eppendorf Services group provides a wide variety of support services such as pipette calibration, instrument maintenance programs, and expedited repair.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.