The following article is the first of a three-part series on imaging large whole mount brain sections for neuroscience research.
The series covers sectioning/staining, scanning, and data management. The next two articles will be published on News-Medical in the coming weeks.
Introduction
Histology, the study of the structure of cells and their formation into tissues and organs, forms the basis of much of our understanding of cell function and disease processes. Histotechnology allows the structure and organization of tissues to be visualized. In addition to facilitating the study of cell biology, this is a powerful tool in the diagnosis of disease.
Brain tissue sample in block form ready for sectioning. Photo credit: Timothy Yates, Indiana University School of Medicine. Image credit: Huron Digital Pathology.
Imaging of tissue samples enables detection and characterization of abnormalities. In a research setting, this furthers understanding of disease processes allowing new treatments to be developed to compensate for, or correct, the abnormality. In clinical practice this facilitates diagnosis and selection of the most appropriate treatment.
Histology of the brain, for obvious reasons, is a research tool rather than a clinical tool. However, this is still invaluable since the brain tissue from donated brains can provide important insights into the underlying pathology of diseases, such as Alzheimer's, which can help in the development of treatments for future patients.
Access to highly detailed histological data is thus fundamental to the advancement of research and treatment outcomes. Acquiring such data requires high-quality histological sections and reliable histological processing. Each step of the collection and preparation process must be performed perfectly in order to obtain valuable information from subsequent imaging.
Mapping the Brain Universe
In neurobiology, obtaining the necessary images can be particularly challenging [Kapelsohn et al. 2015]. Sections for histological imaging are commonly less than one cell thick, yet neurobiological research often requires evaluation of connections between cells and between different areas of the brain.
In order to be able to relate sub-cellular structure to the organization of neural circuitry, large tissue volumes must be visualized at high resolution. With brain sections being as large as 125 mm x 175 mm, the additional size imposes greater challenges to the preparation of perfect tissue sections for imaging. This article highlights the unique challenges of preparing physically large tissue for imaging.
Brain tissue being sectioned using microtome. Photo credit: Timothy Yates, Indiana University School of Medicine. Image credit: Huron Digital Pathology.
Fixation
Fixation is required to preserve the tissue sample in a state as close to that during life as possible. Fixation should thus be carried out as soon as possible after removal of the tissues. A range of fixatives are available and, although none is perfect, formaldehyde is generally considered to be the fixative of choice. Formalin is also known to penetrate tissue well, but does so slowly and this could mean a lengthy wait when using large tissue samples.
Precise initial preparation of any section is critical to ensure good preservation of the sample. It is particularly important that this step is thorough when the tissue volume is large. It is essential to ensure sufficient fluid is used and plenty of time is allowed so that the fixative reaches all parts of the tissue. Maintaining a tissue to fluid ratio of 1:20 is recommended and keep at room temperature.
Rose M. Richardson, Senior Research Technologist, Dr. Bernardino Ghetti’s Lab, Indiana University School of Medicine/Department of Pathology, advises:
“After the brain is sliced at brain cutting and the slabs are submitted for the large sections, allow a day or two more in fixative for the center of the slice to be fixed. Patience will be rewarded.”
Embedding
The preserved tissue is now ready to be embedded in a medium to allow thin slices to be cut cleanly. The medium most commonly used is wax, such as paraffin. Paraffin melts at about 56–58°C and so is safe for embedding neurological tissue.
To prevent the tissue becoming soggy, which will make sectioning difficult, all the water must be removed from the tissue and replaced by the wax. The tissue is progressively dehydrated by immersing it in successively higher concentrations of alcohol.
Another good tip from Ms. Richardson is to “Use ethanol alcohol and not alcohol mixtures with isopropyl which make brain tissue brittle.”
A mixture of 60% ethanol in water is generally good for preserving the shape of cells and preventing buffer salts from the fixative precipitating into the tissue. Only gentle agitation of the sample is required. The solution should then be changed for 80% ethanol, 95% ethanol, and 100% ethanol at around 24-hourly intervals.
Finally, the sample should be transferred into xylene. Wax is soluble in xylene, but not in alcohol, so this step is needed to enable the wax to readily permeate the tissue. At least two changes of xylene should be made to ensure removal of all the alcohol. Furthermore, the xylene step needs to be extended when working with large volume samples to ensure good penetration. It is not adequate to end the xylene step when the tissue has “cleared” and become translucent.
If dehydration is not achieved in the entire tissue, there is a risk that the tissue will separate from the wax during sectioning or will not adhere to the slide.
The tissue can then be transferred to the melted paraffin wax. Two changes of paraffin over 48 hours are recommended for larger tissue samples before embedding. Use of a vacuum oven, if this is possible, will greatly enhance penetration of the wax. The tissue and paraffin can then be transferred to a suitable mold and left to cool and solidify.
Recently, it has become possible to embed up to 40 mouse brain hemispheres or 40 spinal cords into one block using MultiBrain® technology (NeuroScience Associates, Knoxville, Tennessee, USA), whereby speeding up the analysis process. This ensures consistent staining across all sections.
Dr Robert Switzer, President and Chief Scientific Officer of NeuroScience Associates, explained that this technology has now been extended to allow an entire human brain to be embedded "A wide range of sizes of tissue can be accommodated by our technology: mouse spinal cords to human brain hemispheres. Of course, only one human brain hemisphere can be embedded in one block!".
Sectioning
Once cooled, the fixed and processed tissue blocks can be sectioned. When using a microtome to produce large sections, static electricity can be a real problem. Operating a small humidifier close to the sliding microtome and setting up in an area where there are few air currents can help to minimize these unwanted effects.
“A good trick is to have a large ice water bath nearby to float the cut section on until you are ready to transfer to the warm bath for application to the slide”, Ms. Richardson recommends.
The microtome is the most commonly used tool for producing sections, but the vibratome can give better results when cutting thicker sections. Cryostat tissue sectioning is also a widespread method of sectioning brain tissue; however, conventional cryostat sectioning is labor-intensive and associated with a risk of damaging or even losing sections. Tape-assisted cryosectioning significantly improves section quality and the speed of collecting brain sections.

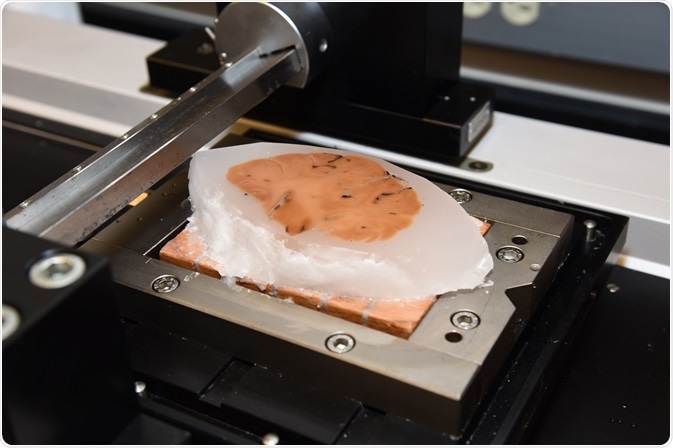
Mounting of thin brain section onto slide. Photo credit: Timothy Yates, Indiana University School of Medicine. Image credit: Huron Digital Pathology.
Mounting
Prior to mounting, the tissue sections should be floated on a warm water bath and the glass slides warmed in an oven. The heating of the section helps ensure that the tissue is smooth and the increased flexibility makes it easier to transfer to the slide. Larger sections are more prone to collapsing and so should be picked up on the glass slide carefully with slow and smooth movements.
Staining
Dyes are used in histology to differentiate features of interest. A range of dyes of differing composition and reactions are available. Knowledge of the properties of a particular dye make it possible to distinguish various tissue structures by dying them different colors.
Unfortunately, dyes are water soluble and so will not stain tissues containing paraffin. It is therefore necessary to remove all the paraffin from the tissue before staining. This can be achieved by repeating in reverse the xylene, alcohol and finally water washes. Again for large tissues, the time taken for the paraffin to be completely removed will be longer than that given in most standard protocols.
Numerous silver staining protocols have been developed, each tailored to reveal specific features. In each case, the designated feature will be stained black. Of particular interest to neurological research is The Campbell-Switzer silver staining method that reveals the neuritic plaques and tangles of Alzheimer's pathology. It is unique in its ability to differentially stain immature and mature plaques, allowing researchers a unique observational capability.
With large tissue blocks, it is more difficult to ensure high-contrast staining throughout the block since the dye has to diffuse though many membranes to reach the center of the block. In neuronal tissue it remains a substantial challenge to achieve consistent high-contrast staining throughout large-volume tissue samples. The situation has recently been improved by the development of large volume en-bloc staining protocols for neuronal tissue [Hua et al 2015].

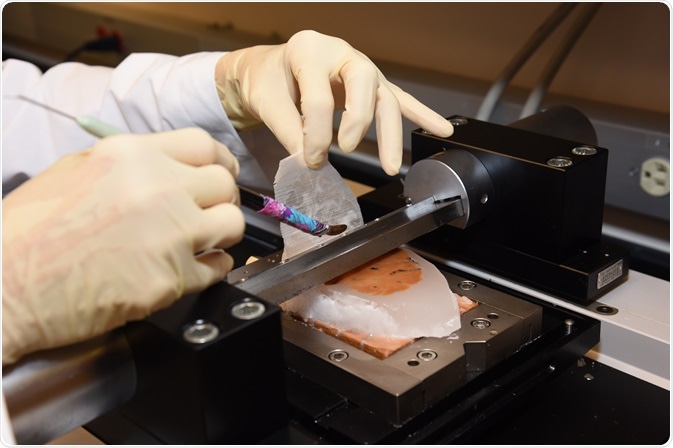
Completed, stained brain section with coverslip. Photo credit: Timothy Yates, Indiana University School of Medicine. Image credit: Huron Digital Pathology.
Coverslipping
The stained section on the slide must then be protected with a coverslip to protect the tissue from being scratched and to preserve the tissue section. It is then necessary to once again remove all the water from the section by subjecting it to the reverse of the process used to remove the paraffin for staining. The stained slide should be passed through a series of alcohol solutions to remove the water, then through clearing agents before the embedding agent is added beneath the glass coverslip.
Since standard slides and coverslips are designed for conventional smaller sections, it will probably be necessary to obtain custom-made slides and coverslips suitable for accommodating the larger sample.
The final slide should be dried overnight at 37–40°C then transferred to a 60°C oven for at least 48 hours to adhere the section.
Summary
Imaging of histology samples has been fundamental to our understanding of how the various systems in the body function and how they malfunction in different disorders.
The value of these images depends on the precise preparation of tissue sections in readiness for scanning. The tissue must be expertly preserved and processed in order to obtain the detailed information needed.
In neurobiological research, it is often necessary to image large-volume brain sections. Such capabilities have been realized relatively recently and allowed valuable insights to be made into the pathophysiology of diseases of the brain, such as Alzheimer's disease. However, such studies still require additional care in the preparation of sections for imaging to ensure quality data are obtained. Furthermore, the larger sections require some lateral thinking in order to find equipment large enough to accommodate the sections since histology is typically performed on a much smaller scale.
This article has provided insight into the extras considerations required when processing large-volume tissue samples. The following articles will discuss the processes of scanning large-volume sections and managing the data obtained.
References
- Hua Y, et al. Nature Communications 2015; volume 6 article 7923. Available at http://www.nature.com/articles/ncomms8923
- Kapelsohn K. Protocol Exchange 2015.Published online 17 March 2015. Available at http://www.nature.com/protocolexchange/protocols/3745#/procedure
- Pinskiy V. et al. PLOS Published online 16 July 2015. Available at http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0102363
About Huron Digital Pathology
 Based in Waterloo, Ontario, Canada, Huron Digital Pathology has a 20 year history designing sophisticated imaging instrumentation. Our end-to-end digital whole slide imaging solutions for digital pathology incorporate our award-winning TissueScope™ digital slide scanners; TissueView™ image viewing, sharing and management platform; and our workflow-enhancing accessories, which include our innovative TissueSnap™ preview scanning station.
Based in Waterloo, Ontario, Canada, Huron Digital Pathology has a 20 year history designing sophisticated imaging instrumentation. Our end-to-end digital whole slide imaging solutions for digital pathology incorporate our award-winning TissueScope™ digital slide scanners; TissueView™ image viewing, sharing and management platform; and our workflow-enhancing accessories, which include our innovative TissueSnap™ preview scanning station.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.