The following article is the second of a three-part series on imaging large whole mount brain sections for neuroscience research.
The first article focused on preparing large tissue sections and the final article on data management in digital imaging will be published on News-Medical in the coming weeks.
Introduction
Imaging stained histological sections allows the microscopic structure and organization of tissues to be visualized. Histotechnology has greatly enhanced our understanding of how the brain works and the disruptions to normal functioning that arise in neurological diseases, such as Alzheimer's disease. Post-mortem studies of sections from donated brains represent the foundation on which our knowledge of both microscopic and macroscopic brain structure and function develops.
In recent years, advances in technology have made it possible to capture pathology images digitally. Having a permanent, searchable record of histology images has tremendous benefits across medical, educational, and research fields. In additional to making cataloguing and retrieval easier, digital images can be readily shared, facilitating collaboration between different research sites. This also opens-up the possibility of remote diagnoses being made by experts, which can be invaluable in the assessment of rare conditions. Furthermore, multiple digital scans can be combined together to create a highly detailed volumetric atlas of the brain.
However, acquiring images of brain sections is particularly challenging since larger sections are commonly needed in order to view the structures of interest. This series highlights special considerations that must be made when imaging large brain sections.
This article highlights some of the key challenges when scanning large brain sections and the means for overcoming them.

TissueScope LE Whole Slide Scanner. Image Credit: Huron Digital Pathology
The importance of digital imaging
As discussed in the first article of this series, brain sections are typically acquired from a sample embedded in a block of paraffin. The sections sliced from the block are transferred to slides for imaging and an appropriate stain applied to facilitate visualisation of the structure of interest. Many slides can thus be derived from a single sample and a variety of stains may be utilised across the different sections. Consequently, although each slide has the potential to provide unique information and has its own identification label, a single slide represents only part of the sample and must be catalogued as such.
The recent development of whole slide imaging allows conventional glass slides to be scanned in order to produce digital slides. Digital slides preserve the slide in its original state and obviate the risk of tissue degradation or fading of stains. Furthermore, the digital slides can be stored electronically allowing easy compilation of all slides from a given sample in logical folders that can be easily searched. Consequently, a digital workflow allows particular slides to be viewed again at a later date to relate them to new findings or to support new conclusions. Furthermore, it provides the image in a format that can be easily shared electronically,
Such digital workflows have the potential to improve accuracy, increase workflow efficiency, and make it possible to share images and integrate them with information systems. Once implemented, such a system results in considerable cost savings providing laboratories with financial returns on their investment.
Digital imaging of the brain has also allowed the creation of three-dimensional reference brains— brain atlases. These comprise digital images of contiguous sections of a healthy or diseased brain along different anatomical planes. Brain atlases can be easily accessed free of charge and provide an invaluable resource for those studying the human brain. As well as showing the surface anatomy, a brain atlas includes maps of the cortex areas with all major structures and subdivisions of the brain labelled. Interactive brain atlases are also available that allow the user to study a particular section of the brain.
Mapping the Brain Universe
Considerations when scanning large brain sections
In order to include all the necessary structures to evaluate neural networks, brain sections can be up to 200 mm x 150 mm. In addition, the thickness of the sections varies considerably and can be as much as 1.5 mm. Scanning such large tissue slides is complex and challenging. In order to obtain high-resolution sub-micron (20x, 40X) scans, the demands on hardware and software requirements are high.
High resolution image scanning technology creates numerous image strips, which are then combined to create the final high resolution image. The strips are created by moving the sample across the lens in one axis while keeping the orthogonal axis stationary. For larger sections, more image strips are created and so more joins are needed. Greater precision is therefore demanded of optomechanical and electromechanical hardware to ensure that the numerous strips are scanned correctly and assembled into a seamless high quality final image with no artifacts. For the largest sections, the scanner must move 200 mm along the x axis and 175 mm along an orthogonal axis (y). The profile of this motion must be extremely precise and is dictated by the smallest resolution of the instrument.
Dr Savvas Damaskinos, CTO, Huron Digital Pathology explained that when scanning larger sections "One of the critical components of a scanning system is the mechanical stage on which the brain tissue slide is mounted for high resolution imaging".

Dr Savvas Damaskinos, Chief Technology Officer, Huron Digital Pathology. Image credit: Huron Digital Pathology.
A further challenge when scanning large tissue is the duration of the scanning process, which is considerably longer than for conventional sections. Technologies for imaging smaller tissues are fast, but do not accommodate larger sections. Although solutions for imaging larger tissue have been developed, sometimes it can take a long time for the imaging to be undertaken, requiring considerable investment of time. It is therefore important to ensure that the stage velocity motion performance is maintained at a scan speed suitable for imaging a large tissue slide within a practical timescale. Failure to produce a high-quality in-focus image can significantly slow down scan throughput. The velocity error of the scanning axis is dependent on the scan frequency and exposure time of the camera.
Dr Damaskinos emphasized that when imaging larger tissue "Maintaining a high constant velocity so as to avoid overwhelming the frame rate of the camera and to avoid pixel smearing is critical to producing a high quality image".
The vast size differential between conventional sections and large brain sections introduces the problem of securing the slide in the scanner without introducing potential for damaging the slide or risking suboptimal scanning by interfering with the optical path arrangement. In addition, once the slide is in the scanner, it is necessary to identify the region of interest, the slope and focus dots, and determine a location to place a white balance box. This requires provision of a rapid low resolution preview image before a high-resolution scan of the selected region of interest is performed. With large samples, it is particularly challenging to gain the necessary scan parameters without the investment of lots of time.
Dr Damaskinos commented "The challenge is to design optical paths to perform fast preview imaging of large areas, usually in brightfield, that is of sufficient quality to set up and perform a high-resolution scan".
In addition to their size, the very nature of large tissue samples can also hinder imaging. Typically, standard sections have a relatively uniform thickness of around 5 um. In contrast, there can be considerable variability in the thickness of large sections. Consequently, larger sections require a more complex workflow, e.g. more advanced outlier focus rejection algorithms, in order to obtain the focus surface map needed to achieve a high-quality image. In addition, the thickness of large format slides can exceed the depth of field of the optics by several orders. Again, this necessitates a more complex scan setup and scan acquisition workflow to achieve an accurate focus map that will produce a high-quality image.
The greater complexity of large tissue imaging also has a significant impact on software capabilities and availability of computing memory space. A JPEG compressed image of a large brain slide tissue section at 20x resolution will be at least 100 GB, which is 10‑100 times larger than comparable images of a standard slide.
Dr Damaskinos advises that "Intelligent software error handling and automatic recovery capability is critical for large format tissue scans to reduce scan failures."
TissueScope™ scanner
Huron’s TissueScope™ (Huron Digital Pathology, Waterloo, Ontario, Canada) scanner has been developed to address the challenges of large tissue scanning. It is capable of digitizing a variety of slides ranging from regular 75 mm × 25 mm glass slides to large glass slides of 200 mm × 150 mm. This is achieved using slide holders that fit the fixed stage of the scanner but accommodate different fittings to accommodate and safely scan a wide range of format slides—from standard to whole mount. Once the slide is in the slide holder, the operator does not need to touch the slide again and the scanner only handles the slide holder - this minimizes the potential of slide damage.

TissueScope LE120 Whole Slide Scanner. Image Credit: Huron Digital Pathology
The TissueScope Scanner also allows for rapid initial scanning so that the region of interest and required scan parameters can be quickly determined before the full high-resolution scan is initiated. It achieves this by including two independent optical paths; one that images a 200 mm x 150 mm area in less than 10 seconds and automatically determines all scan parameters, and the second that performs a high-resolution scan.
In addition, the TissueSnap™ device increases the scan speed by acquiring fast preview images of a slide and transferring the scan parameters to the scanner for the high-resolution imaging. While the TissueScope LE120 is scanning, the TissueSnap can capture preview images of slides yet to be scanned. The preview images are sent instantly to the main scanner’s user interface. As the scanner continues to acquire high-resolution images, the next batch of slides can be previewed and setup simultaneously without interruption.
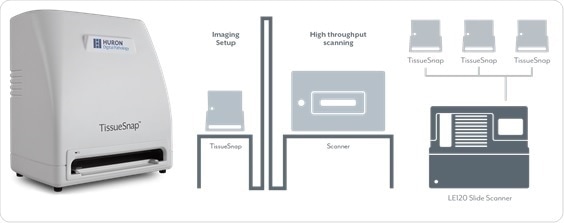
TissueSnap Preview Station. Image Credit: Huron Digital Pathology
A resolution of at least 0.25 um pixel (as required to achieve the 40X resolution needed in digital pathology) is achieved in the TissueScope scanner. Furthermore, the scanner maintains motion performance at a scan speed suitable for imaging a large tissue slide in a reasonable time. Including setup time, imaging a 175 mm x 125mm human brain section at 20X will take 35 minutes and at 40X, 65 minutes.
Summary
Histology has always been a key component of research programs. With the development of digital imaging it has become an even more valuable tool. The ability to obtain permanent images of a section that can be readily catalogued and shared has significantly enhanced the utility of histotechnology.
Furthermore, the availability of automated systems that can produce high-quality, high-resolution images has made histology work flow more efficient. leading to considerable cost savings.
References
About Huron Digital Pathology
 Based in Waterloo, Ontario, Canada, Huron Digital Pathology has a 20 year history designing sophisticated imaging instrumentation. Our end-to-end digital whole slide imaging solutions for digital pathology incorporate our award-winning TissueScope™ digital slide scanners; TissueView™ image viewing, sharing and management platform; and our workflow-enhancing accessories, which include our innovative TissueSnap™ preview scanning station.
Based in Waterloo, Ontario, Canada, Huron Digital Pathology has a 20 year history designing sophisticated imaging instrumentation. Our end-to-end digital whole slide imaging solutions for digital pathology incorporate our award-winning TissueScope™ digital slide scanners; TissueView™ image viewing, sharing and management platform; and our workflow-enhancing accessories, which include our innovative TissueSnap™ preview scanning station.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.