Single-cell sequencing includes the isolation of a single cell and studying its sequence information with the latest sequencing technologies to fully characterize cells and gain invaluable insight into cellular differences.
The single-cell genomics field is experiencing rapid advancement, despite researchers within the field encountering numerous challenges, such as sensitivity, reproducibility, scalability, and cost, especially when vast numbers of cells are being analyzed.
However, miniaturization and automation have been shown to address these limitations.
Nextera XT sample prep kits (manufactured by Illumina, Inc., USA) are often utilized for the preparation of DNA libraries based on enzymatic fragmentation of DNA samples with only very small volumes of input DNA used.
To guarantee high precision and accuracy, most library preparation protocols suggest using volumes within the range of large-volume liquid handlers or manual pipettes. However, only a small portion of these preparations is needed for sequencing.
The use of a liquid handler that handles low volumes of solutions with various viscosities enables the preparation of smaller reaction volumes, which saves on both the cost of reagent and sample input.
This article describes a high-throughput, miniaturized workflow which involves the study of differentiated pancreatic stem cells with single-cell RNA-seq (scRNA-seq) in Dr. Louise Laurent’s laboratory at the University of California, San Diego, United States.
Key benefits
Mosquito liquid handlers for low-volume single-cell RNAseq library preps:
- Reduce cost through the miniaturization of reagent volumes
- Provide reliable pipetting of solutions with low to high viscosity
- Increase reproducibility and data quality with accurate pipetting at low volumes
Miniaturization of the library prep volumes was achieved using SPT Labtech’s mosquito LV and HV liquid handlers, which accurately dispense volumes of between 25 nL and 1.2 μL and 0.5 to 5 μL, respectively.
These liquid handlers utilize true positive-displacement pipetting technology, meaning they can operate at the same setting across liquids with various viscosities, including buffers, alcohols, enzymes, and low to high concentrations of gDNA (200-300 ng/μL).
The resultant single-cell RNA-seq data was analyzed to establish the reproducibility of this system and its capability for classifying cells at different stages of differentiation as well as individual cells within each stage.
The following journal publication includes more detailed results and discussion: J Lab Autom. 2016 Aug; 21(4):557-67.
Case study: Single-cell analysis of differentiated human pancreatic stem cells methods
Human embryonic stem cells (WA09) were differentiated from pancreatic progenitor cells and then subsequently dissociated into a cell suspension.
An average cell concentration of 2.5 x 105 cells/mL was loaded into the C1 Single-Cell Auto Prep System (Fluidigm, San Francisco, United States), which generated and amplified cDNA from cells at two differentiation stages.

Image Credit: SPT Labtech
Following this, two cells from stage 1 and two cells from stage 2 were chosen to undergo single-cell analysis.
The resultant cDNA was diluted to a final concentration of 0.1 ng/µL. Following this, it was converted to Illumina sequencing libraries utilizing both the Nextera XT kit (Illumina, San Diego, United States) and the mosquito LV liquid handler, as displayed in Figure 1.
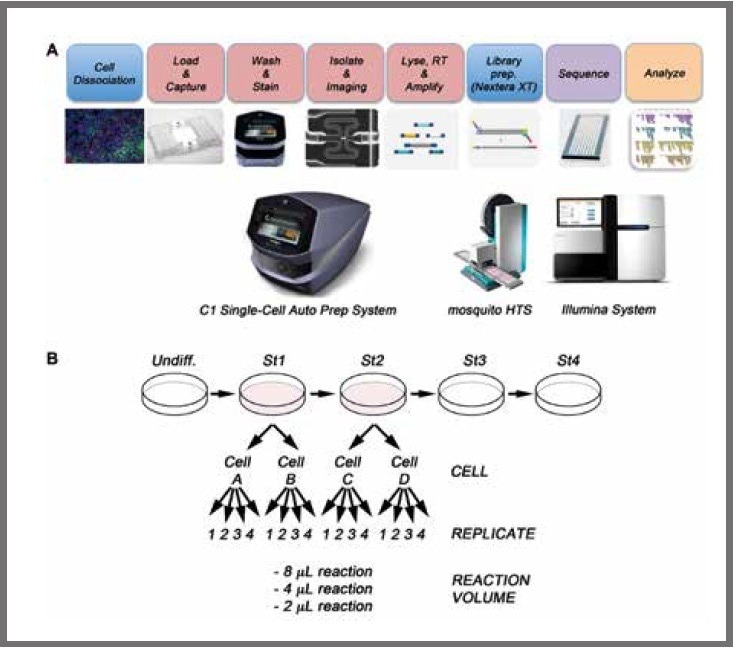
Figure 1. (a) Schematic of workflow for the single-cell sequencing of differentiated pancreatic stem cells. After differentiation, the cell cultures are dissociated to single cell suspensions and loaded onto a Fluidigm C1 Single‑Cell Auto Prep Array for mRNA-Seq where the mRNA is reverse transcribed and the cDNA amplified. Libraries are prepared using the Nextera XT kit and mosquito LV (SPT). Finally, libraries are pooled and sequenced on an Illumina HiSeq 2500. (b) WA09 human embryonic stem cells were differentiated in vitro to the pancreatic lineage. Two independent cells from stage 1 (cell A and cell B) and two cells from stage 2 (cell C and cell D) were selected for single-cell analysis. For library preparation, 2 μL, 4 μL, and 8 μL final volume reactions were tested, with four technical replicates per reaction volume. Image Credit: SPT Labtech
Libraries were produced in three distinct final reaction volumes of 2 µL, 4 µL, and 8 µL in quadruplicate in 384-well PCR plates utilizing as little as 20, 40, and 80 pg of cDNA per reaction, respectively. When it comes to volumes and sample input, Illumina advises that 50 µL and 1 ng are used, respectively.
Excess primer dimers, salts, nucleotides, and enzymes were removed using the process of magnetic bead clean-up.
High-throughput and low-volume bead clean-up was carried out using the SPT Labtech mosquito HV liquid handler equipped with Agencourt AMPure XP magnetic beads (supplied by Beckman Coulter, Brea, USA) and a 384 well magnet (supplied by SPT Labtech, SZZ00136).
A total of 1.8 µL of beads and 2 µL from each single-cell library were mixed, and the libraries were cleaned up utilizing the standard protocol to pull down and wash the beads, and the libraries were then eluted from the beads.
Analysis was conducted on the resultant purified libraries using the BioAnalyzer (Agilent Technologies, Santa Clara, USA), and normalized to 0.1 ng/µL, and then pooled and loaded onto a sequencer (HiSeq 2500, Illumina).
With a total of 48 pooled libraries, they were then sequenced at an average total read depth of 5.6 million reads per sample.
The sequencing data attained from this was analyzed, and correlations between the three distinct reaction volumes and technical and biological replicates were examined for reproducibility and quality.
Results
Miniaturization does not impact the technical replicates
The Nextera XT kit has been validated by Fluidigm for a final reaction volume down to 10 µL, with between 125-375 pg of sample input. Using mosquito LV, the sample input and reaction volume were decreased to 20 pg and 2 µL, respectively.
After DEseq normalization of the data set, correlations between replicates, reaction volumes and cell types were calculated using Pearson’s correlation.
The mean correlation coefficient was found to be higher than 0.936 between each technical replicate at each reaction volume for each cell, both with and without down-sampling. The correlation coefficients between the various reaction volumes for a given cell were all higher than 0.918.
To further determine whether the reaction volume affected the reproducibility of the library preparation, the coefficient of variation (CV) for each library was calculated using DESeq normalized data.
The CVs were found to be from 2.9 to 3.8 for all cells and all reaction volumes.
When the mean CV for each cell, irrespective of reaction volume, was calculated, there were no significant differences between the overall CVs and the CVs from each reaction volume separately (see Figure 2).
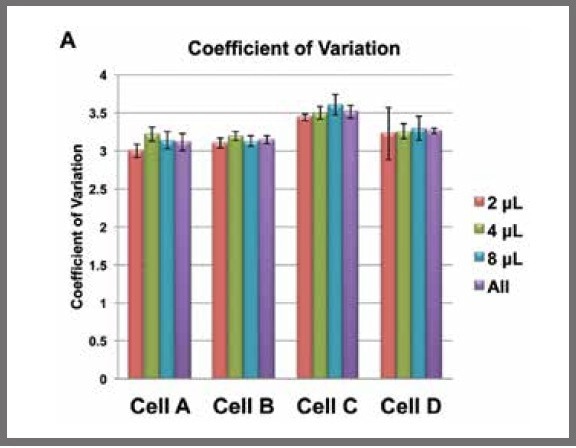
Figure 2. Mean coefficients of variation (CVs) for each reaction volume for each cell calculated from DESeq normalized data. The purple bars are the mean CVs for each cell, irrespective of reaction volume. Image Credit: SPT Labtech
Table 1. Volumes (nl) of reagents and cDNA pipetted by mosquito LV to obtain total reaction volumes of 2, 4 or 8 mu;L. Source: SPT Labtech
| reagent volume (nL) |
reaction volumes (μL) |
| 2 |
4 |
8 |
| Atm enzyme mix |
200 |
400 |
800 |
| TD buffer |
400 |
800 |
1,600* |
| cDNA (0.1 ng/L) |
200 |
400 |
800 |
| NT buffer |
200 |
400 |
800 |
| NPM enzyme mix |
600 |
1,200* |
2,400* |
| Double Index |
200
200 |
400
400 |
800
800 |
* total volume was made up of multiple pipetting of smaller volumes
Miniaturization does not impact the biological replicates
Two distinct clustering methods were employed, namely 2D principal component analysis (displayed in Figure 3a) and hierarchical clustering (displayed in Figure 3b).
These revealed a clear separation between the libraries from each of the four cells. Notably, the libraries did not cluster by reaction volume, even within a single cell.
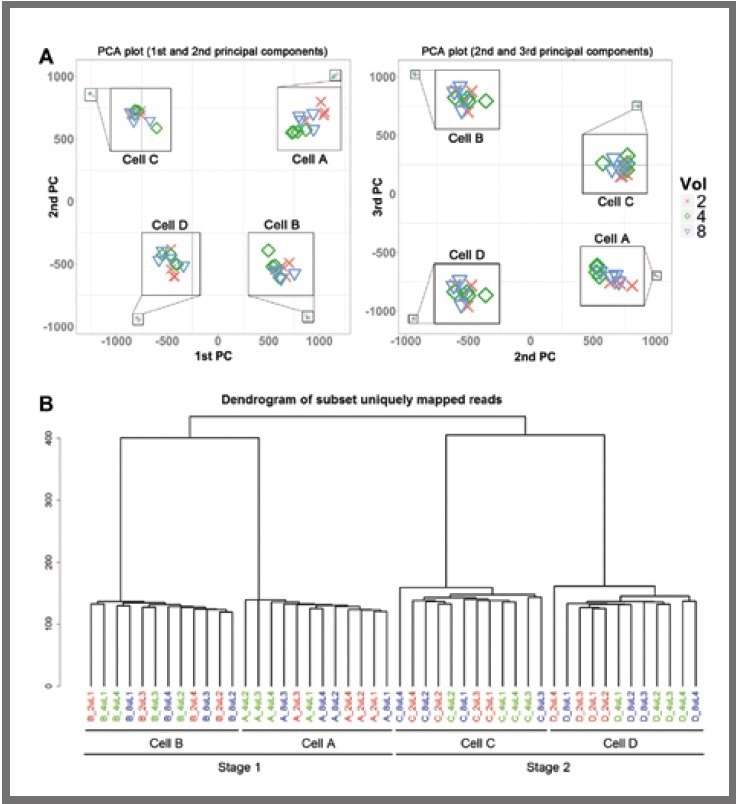
Figure 3. Clustering analysis. (a) Principal component analysis (PCA) for libraries. (b) Hierarchical clustering of all. Image Credit: SPT Labtech
Miniaturization does not impact library complexity
A possible issue regarding the decreasing reaction volume for library preparation is the introduction of sampling error. This may lead to reduced detection of transcripts expressed at low levels, reducing the complexity of the libraries.
Firstly, the union of overlapping transcripts among the four replicates for each cell for each reaction volume was obtained, and subsequently, the overlaps between the 2-μL, 4-μL, and 8-μL libraries were examined, as presented in Figure 4 (top).
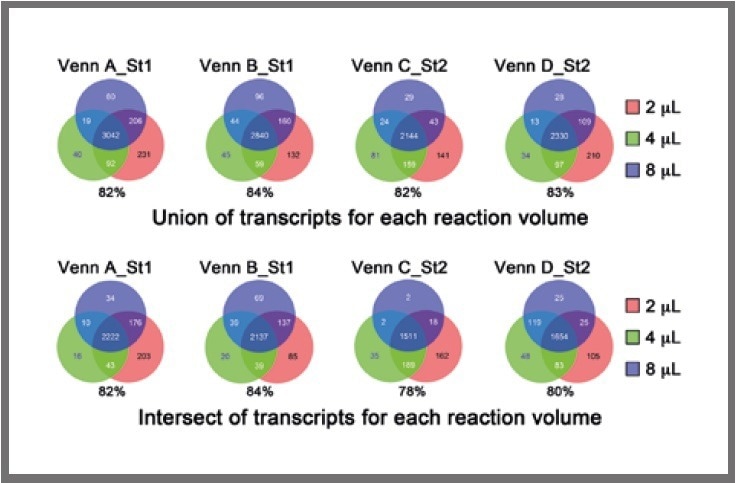
Figure 4. Venn diagrams displaying the overlap in detected transcripts among the different reaction volumes, using the union (top) or intersect (bottom) of detectable genes in the four replicates. The percentage of transcripts in the common region of intersection (i.e., 2 μL, 4 μL , 8 μL) compared with all transcripts (i.e., 2 μL, 4 μL, 8 μL) is shown below each Venn diagram. Image Credit: SPT Labtech
Following this, the intersection of overlapping transcripts among the four replicates for each cell for each reaction volume was obtained, and the overlaps between the 2-μL, 4-μL, and 8-μL libraries were examined, as presented in Figure 4 (bottom).
In general, the overlaps were highly similar across volume ranges, verifying that miniaturization does not impact the complexity of the libraries.
Conclusions
The subsequent single-cell RNA-seq data revealed that miniaturization carried out with the use of mosquito low-volume liquid handlers does not impact the complexity of the prepared library or the reproducibility whilst delivering substantial cost savings via miniaturization of reaction volumes.
Even at low reaction volumes, it was possible to distinguish between cells at different stages of differentiation and also between individual cells within each stage.
This technical advancement will significantly reduce both the labor and cost involved in single-cell transcriptome studies, allowing for hundreds to thousands of single cells to be analyzed.
Acknowledgments
Produced from materials originally authored by SPT Labtech. The original authors wish to thank Dr. Louise Laurent’s group at Sanford Consortium of Regenerative Medicine, University of California, San Diego (UCSD), United States, for their collaboration.
About SPT Labtech
We Design and Manufacture Robust, Reliable and Easy-to-Use Solutions for Life Science
We enable life scientists through collaboration, deep application knowledge, and leading engineering to accelerate research and make a difference together. We offer a portfolio of products within sample management, liquid handling, and multiplexed detection that minimize assay volumes, reduce material handling costs and put the discovery tools back in the hands of the scientist.
At the Heart of What We Do
Many of our innovations have been born out of the desire to create solutions to existing customer problems; and it’s this ethos that drives SPT Labtech’s R&D efforts. Our strengths come from the trust our customers have with us to develop truly unique, automated technologies to meet their needs. We combine cutting edge science with first-rate engineering to put customers at the heart of everything we do.
A Problem-Solving State of Mind
The substantial breadth of expertise within our company enables us to be involved in the full life cycle of our products from the initial design concept, mechanical and software engineering and prototyping, to final manufacture and sale. These qualities allow us to offer the best possible technical and mechanical support to all the equipment that we supply, hence maintaining excellent client relationships.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.