Tuberculosis (TB) is a significant global health challenge caused by Mycobacterium tuberculosis. Nontuberculous mycobacteria (NTM) can produce symptoms similar to TB, with some species leading to serious pulmonary issues.
Image Credit: Kateryna Kon/Shutterstock.com
Accurate detection is essential to prevent misdiagnosis and ensure the right treatment, particularly in cases involving drug-resistant TB or NTM infections. The Hi-PCR® MTB/NTM Probe PCR Kit offers a quick and dependable method for detecting and differentiating MTB and NTM in clinical samples. Utilizing real-time PCR technology with specialized hydrolysis probes, this kit delivers results in under 70 minutes, making it an invaluable tool for clinical diagnostics, monitoring, and research.
Introduction
Tuberculosis (TB) continues to be a major global health issue, resulting in millions of new cases and fatalities annually. It is primarily caused by the Mycobacterium tuberculosis (MTB) complex and predominantly affects the lungs. Meanwhile, nontuberculous mycobacteria (NTM) can also mimic TB symptoms, with some species being notably significant in pulmonary disease.
Conventional TB diagnostic techniques, such as smear microscopy and culture, are often slow and lack sensitivity and specificity in distinguishing between TB and NTM, leading to delays in both diagnosis and treatment. To overcome these obstacles, probe-based real-time PCR assays have been created to accurately identify and differentiate MTB and NTM infections.
The Hi-PCR® MTB/NTM Probe PCR Kit provides a fast and reliable approach for the simultaneous detection of MTB and NTM in clinical samples, facilitating timely and appropriate treatment decisions.
Overview of the technology that underpins the Hi-PCR® MTB/NTM Probe PCR Kit
The Hi-PCR® MTB/NTM Probe PCR Kit employs real-time PCR technology to amplify bacterial DNA from clinical specimens, followed by detection with fluorescent probes. This approach enables quick and precise detection and differentiation of Mycobacterium tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM) in a single-tube assay.
The method involves amplifying target DNA using hydrolysis probes, which are short oligonucleotides with a fluorescent dye attached at the 5' end and a quencher dye at the 3' end. During amplification, the probe is cleaved, separating the dye from the quencher, leading to increased fluorescence that is monitored in real-time. This allows for the immediate and accurate identification of MTB and NTM DNA.
The Hi-PCR® MTB/NTM Probe PCR Kit includes a single tube containing oligonucleotides (primers and probes) designed to amplify conserved regions of MTB and NTM genes, along with an internal control amplification system.
Each component is labeled with different fluorophores: the MTB complex target is detected in the FAM channel, the NTM target in the ROX channel, and the internal control target in the JOE channel.
Work flow
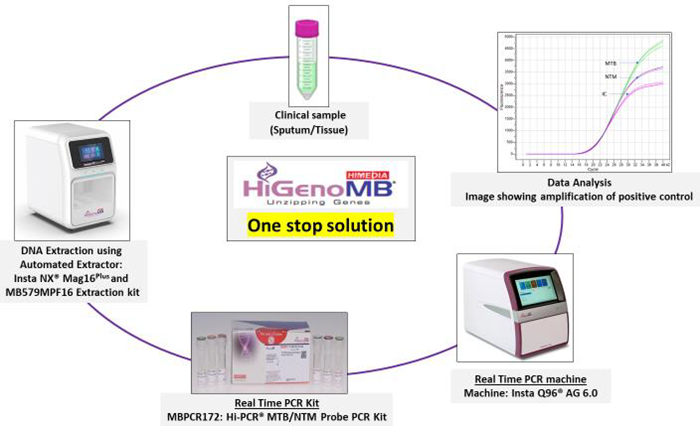
Figure 1. Image representing workflow of the process and One Stop Solution offered by HiGenoMB®. Image Credit: Himedia Laboratories Private Limited
Molecular features
- Simultaneous detection of MTB and NTM in a single assay
- High sensitivity – 1 copy per microliter (μl) for MTB; 10 copies per μl for NTM
- High specificity – No cross-reactivity with pathogens having a similar clinical presentation
Technology features
- Rapid and reliable results within 70 minutes
- Includes all reagents & controls for validity of the test
- Open system – Compatible with 4-channel and 5-channel qPCR cyclers
- Wet-lab assays validated on the Bio-Rad CFX Opus 96, Applied Biosystems Quant Studio 5 and Insta Q96® Plus Real Time PCR Systems
Applications
- Clinical Diagnostics
- Epidemiological Surveillance
Performance validation
The Hi‐PCR®MTB/NTM Probe PCR Kit has been rigorously validated across multiple platforms, including Bio-Rad CFX Opus 96, Applied Biosystems Quant Studio 5, and Insta Q96® Plus Real-Time PCR Systems.
Analytical sensitivity
Limit of Detection (LoD)
The Limit of Detection (LoD) is defined as the concentration (copies per μl of the eluate) of a target molecule that can be detected with 95% probability, in accordance with CLSI EP17-A2 guidelines.
The LoD assay for the Hi-PCR® MTB/NTM Probe PCR Kit was conducted using 20 replicates on the Biorad CFX Opus 96, Applied Biosystems Quant Studio 5, and Insta Q96® Plus Real-Time PCR Systems.
The assay utilized ATCC standards of Quantitative Genomic DNA from Mycobacterium tuberculosis strain H37Ra (ATCC 25177DQ) and Mycobacterium avium subsp. paratuberculosis strain K-10 (ATCC BAA-968D). The detectable limit for the Hi-PCR® MTB/NTM Probe PCR Kit was found to be 1 copy per μL for MTB and ≤ 10 copies per μL for NTM.
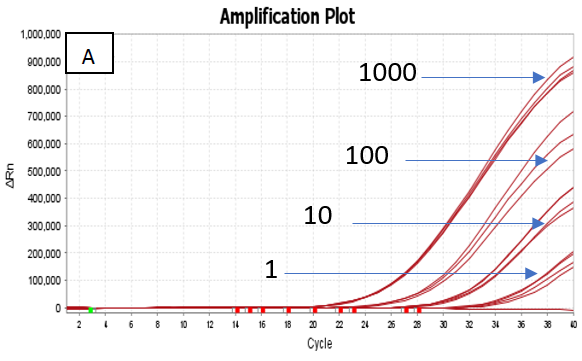
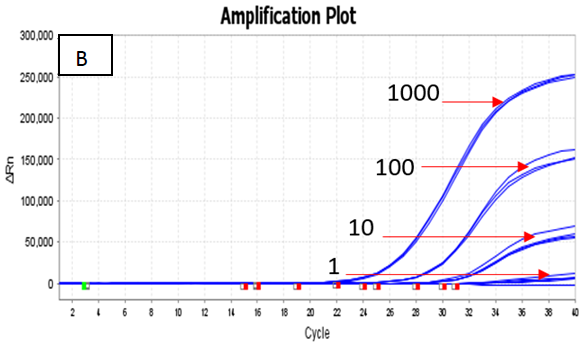
Figure 2. Dilution series (1000 copies/μl, 100 copies/μl, 10 copies/μl and 1 copy/ μl) of A) Quantitative Genomic DNA from Mycobacterium tuberculosis strain H37Ra (ATCC 25177DQ) and B) Quantitative Genomic DNA from Mycobacterium avium subsp. paratuberculosis strain K‐10 (ATCC BAA‐968D) run on the Applied Biosystems Quant Studio 5 Real Time PCR Systems. Image Credit: Himedia Laboratories Private Limited
Analytical specificity
Inclusivity – In silico
The analytical specificity of the Hi-PCR® MTB/NTM Probe PCR kit was ensured by in-silico analysis of the oligonucleotides (primers and probes). The oligonucleotide sequences of all the targets were checked by sequence comparison against the following sequences of MTB complex and NTM strains available in the GenBank database and specificity was found to be 100%.
Source: Himedia Laboratories Private Limited
| MTB |
| Mycobacterium tuberculosis H37Ra, H37Rv, H37RvSiena, (Zopf 1883) Lehmann and Neumann 1896, Mycobacterium tuberculosis complex sp. N0072, Mycobacterium tuberculosis strain 1-19, 2.2.1, H-19-0008, H-20-0024, N1015 |
| Mycobacterium bovis AF2122/97, BCG str. Tokyo 172, BCG strain Moreau PL, BCG SL 222 Sofia, BCG strain Russia 368, BCG str. Moreau RDJ, BCG str. Tokyo 172 |
| Mycobacterium africanum strain 25, UT307, GM041182, |
| Mycobacterium microti strain 12, OV254, Reed 1957 |
| Mycobacterium caprae strain Allgaeu |
| Mycobacterium canetti strain ET1291 |
| Mycobacterium orygis strain NIAB_BDWBCSHFL_1, strain MUHC/MB/EPTB/Orygis/51145 |
| NTM |
| Mycobacterium avium complex (MAC), M. abscessus, M. kansasii, M. intracellulare, M. scrofulaceum, M. fortuitum complex, M. malmoense , M. interjectum, M. gordonae, M. flavescens, M. chelonae, M. simiae, M. gastri, M. smegmatis, M. avium, M. celatum, M. terrae complex, M. xenopi, M. marinum, M. phlei, M. vaccae, M. ulcerans, M. tusciae, M. triplex, M. septicum, M. mucogenicum, M. asiaticum, M. intermedium, M. chimaera, M. senegalense, M. parascrofulaceum, M. toakiense, M. haemophilum, M. aurum, M. thermoresistable, M. aichiense, M. thermophilum, M. neoaurum, M. kubicae, M. bohemicum, M. shimoidei, M. rhodesia, M. florentinum, M. hiberniae, M. mucogenicum, M. colombiense, M. wolinsky , M. longobardum, M. nonchromogenicum |
Analytical reactivity
The analytical reactivity of the Hi-PCR® MTB/NTM Probe PCR Kit was confirmed through wet lab testing of the oligonucleotides (primers and probes) against commercial controls for MTB and NTM.
This testing included Quantitative Genomic DNA from Mycobacterium tuberculosis strain H37Ra (ATCC 25177DQ), Quantitative Genomic DNA from Mycobacterium tuberculosis variant bovis BCG strain TMC 1011 (ATCC 35734D), and Quantitative Genomic DNA from Mycobacterium avium subsp. paratuberculosis strain K-10 (ATCC BAA-968D).
Additionally, it involved Genomic DNA from Mycobacterium abscessus strain L948 (ATCC 19977D-5), Quantitative Genomic DNA from Mycobacterium microti (ATCC 19422DQ), Genomic DNA from Mycobacterium gordonae strain TMC 1327 (ATCC 35760D-5), Genomic DNA from Mycobacterium marinum strain M (ATCC BAA-535D-5), and Genomic DNA from Mycobacterium smegmatis strain mc(2)155 (ATCC 700084D-5).
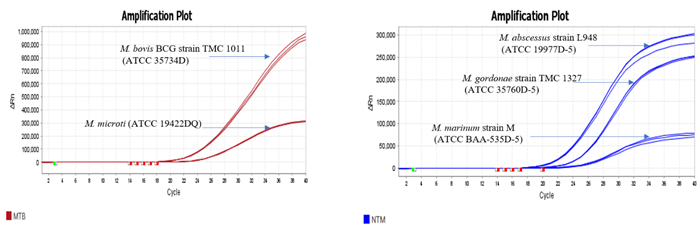
Figure 3. Image representing probe based Real-Time amplification plots of ATCC standards (MTB and NTM) confirmed by Hi-PCR® MTB/NTM Probe PCR kit on Applied Biosystems Quant Studio 5 Real Time PCR System. Image Credit: Himedia Laboratories Private Limited
Cross-reactivity and interference with other microorganisms
Wet testing was conducted against commercial genomic or synthetic DNA/RNA of the pathogens listed in the following table, using the Applied Biosystems Quant Studio 5, to assess any potential cross-reactivity. None of the tested pathogens displayed reactivity to the primers and probes of the Hi-PCR® MTB/NTM Probe PCR Kit.
Source: Himedia Laboratories Private Limited
| |
|
| Influenza A virus (H3N2) strain A/Wisconsin/15/2009 (VR-1882DQ) |
Corynebacterium diphtheriae strain NCTC 13129 (700971D-5) |
| Human coronavirus 229E (ATCC VR-740DQ) |
Haemophilus influenzae (51907DQ) |
| Human metapneumovirus hMPV RNA (ATCC VR-3250SD) |
Pseudomonas aeruginosa strain PAO1-LAC (47085DQ) |
| Enterovirus 68 strain Fermon (ATCC VR-1826) |
Staphylococcus aureus subsp. aureus (43300DQ) |
| Human parainfluenza virus 1 strain C35 (ATCC VR-94DQ) |
Chlamydophila pneumoniae strain CM-1 (1360DQ) |
| Human parainfluenza virus 3 strain C 243 (ATCC VR-93DQ) |
Mycoplasma pneumoniae strain M129-B7 (29342DQ) |
| Human respiratory syncytial virus strain 18537 (ATCC VR-1580DQ) |
Legionella pneumophila subsp. pneumophila (33152DQ) |
| Influenza B virus (ATCC VR-1804DQ) |
Bordetella pertussis (9797DQ) |
| Influenza A virus (H1N1) strain A/PR/8/34 (ATCC VR-1469DQ) |
Candida albicans strain SC5314 (MYA-2876DQ) |
| Human coronavirus NL63 RNA (ATCC VR-3263SD) |
Aspergillus niger strain A1144 3528.7 (1015DQ) |
| Measles virus strain Edmonston (VR-24D) |
Aspergillus flavus strain SN 3 (9643DQ) |
| Human adenovirus 1 strain Adenoid 71 (VR-1DQ) |
Streptococcus pyogenes strain Bruno (19615DQ) |
| Human parainfluenza virus 2 strain Greer (VR-92DQ) |
|
Cross-reactivity analysis – In silico
The sequences of the oligonucleotides (primers and probes) utilized in the Hi-PCR® MTB/NTM Probe PCR Kit underwent BLAST (Basic Local Alignment Search Tool) analysis against the organisms shown in the table below. The in-silico analysis revealed no significant cross-reactivity for any of the evaluated sequences.
Source: Himedia Laboratories Private Limited
| |
|
| Epstein Barr virus (taxid:10376) |
Klebsiella pneumoniae (taxid:573) |
| Human bocavirus (taxid:329641) |
Streptococcus pyogenes (taxid:1314) |
| Rhinoviruses (taxid:12059) |
Streptococcus group G (taxid:1320) |
| Cytomegalovirus (taxid:10358) |
Escherchia coli (taxid:562) |
| VZV (taxid:10335) |
Moraxella catarrhalis (taxid:480) |
| Herpes simplex virus 1 (taxid:10298) |
Haemophilus parainfluenzae (taxid:729) |
| Herpes simplex virus 2 (taxid:10310) |
Corynebacterium diphtheriae (taxid:1717) |
| Severe acute respiratory syndrome coronavirus (taxid:694009) |
Corynebacterium ulcerans (taxid:65058) |
| MERS-CoV (taxid:1335626) |
Salmonella (taxid:590) |
| Human Parainfluenza Virus-4 (taxid:2560526) |
Bordetella pertussis (taxid:520) |
| Measles morbillivirus (taxid:11234) |
Legionella pneumophila (taxid:446) |
| SARS-CoV-2 (taxid:2697049) |
|
The precision of the Hi-PCR MTB/NTM Probe PCR Kit was evaluated across various conditions, including:
- Intra-assay variability: Variability within a single experiment.
- Inter-assay variability: Variability between different experiments.
- Inter-day variability: Variability across three different days.
- Inter-operator variability: Variability among three different operators.
- Inter-instrument variability: Variability between three different PCR thermal cyclers.
- Inter-lot variability: Variability between three different lots of the kit.
Variability data were assessed in terms of standard deviation (SD) and coefficient of variation (CV) based on threshold cycle (Ct) values. Total variability was determined by combining results from all five types of variability analyses.
Table 3. Data showing variability observed from Precision (repeatability and reproducibility) studies. Source: Himedia Laboratories Private Limited
| Repeatability and Reproducibility data |
| Variability |
Mean (Ct) |
MTB |
NTM |
IC |
| Intra-assay |
SD |
0.133 |
0.19 |
0.09 |
| %CV |
0.53 |
0.74 |
0.34 |
| Mean (Ct) |
24.82 |
24.76 |
26.23 |
| Inter-assay |
SD |
0.71 |
0.45 |
0.19 |
| %CV |
2.94 |
1.87 |
0.72 |
| Mean (Ct) |
24.11 |
24.0 |
26.12 |
| Inter-Day |
SD |
0.79 |
0.46 |
0.05 |
| %CV |
3.26 |
1.91 |
0.2 |
| Mean (Ct) |
24.16 |
24.04 |
26.23 |
| Inter-Operator |
SD |
0.057 |
0.004 |
0.04 |
| %CV |
0.22 |
0.0 |
0.15 |
| Mean (Ct) |
24.88 |
24.76 |
26.24 |
| Inter-Instrument |
SD |
3.22 |
0.36 |
1.45 |
| %CV |
14.06 |
1.45 |
7.48 |
| Mean (Ct) |
22.89 |
24.76 |
24.57 |
| Inter-lot |
SD |
0.18 |
0.06 |
0.69 |
| %CV |
0.74 |
0.24 |
0.03 |
| Mean (Ct) |
24.41 |
24.74 |
26.62 |
| Total variability |
SD |
0.76 |
0.38 |
26.0 |
| %CV |
3.12 |
1.54 |
0.72 |
| Mean (Ct) |
24.33 |
24.51 |
26 |
Clinical performance evaluation
The performance of the Hi-PCR MTB/NTM Probe PCR Kit was evaluated using clinical specimens to confirm aspects of assay performance. A total of 40 clinical samples were assessed and compared with a Reference CE-IVD PCR Multiplex Kit. Bacterial nucleic acids were extracted using the HiPurA® Pre-filled Plate for MTB DNA Purification (Cat no: MB579MPF16). Results for MTB are illustrated in the table below.
Table 4. Table representing Sensitivity, Specificity, PPV and NPV data of the Hi-PCR® MTB/NTM Probe PCR Kit. Source: Himedia Laboratories Private Limited
| |
Value |
95%CI |
| Diagnostic sensitivity |
100% |
81.5-100% |
| Diagnostic specificity |
100% |
78.1-100% |
| Positive Predictive Value |
100% |
81.5-100% |
| Negative Predictive Value |
100% |
78.1-100% |
Case study
DNA extracted from an MTB-positive sample was serially diluted at ratios ranging from 1:10 to 1:1000 to simulate samples with low target concentrations. These diluted samples were tested using the Hi-PCR® MTB/NTM Probe PCR Kit. The results showed that even samples with low concentrations of the target gene were successfully detected as positive, with the generated Ct values remaining within the cutoff threshold for detection.
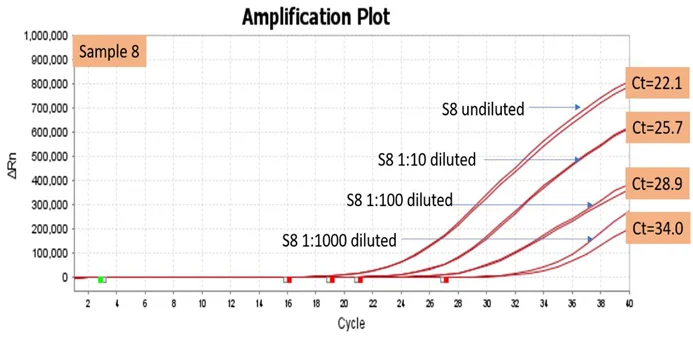
Figure 4. Image showing amplification of Sample 8 and its dilution series (1:10-1:1000) with the corresponding Ct values confirmed by Hi-PCR® MTB/NTM Probe PCR kit on Applied Biosystems Quant Studio 5 Real Time PCR System. Image Credit: Himedia Laboratories Private Limited
Conclusion
The Hi-PCR MTB/NTM Probe PCR Kit is a highly effective tool for the rapid and accurate identification and differentiation of Mycobacterium tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM).
Its exceptional sensitivity, specificity, and user-friendliness make it an indispensable resource for healthcare providers, public health officials, and researchers in combating tuberculosis and related infections. By facilitating prompt diagnoses and informed decision-making, this kit plays a crucial role in managing outbreaks, improving patient outcomes, and supporting global efforts to control and prevent tuberculosis.
About Himedia Laboratories Private Limited
With a presence in more than 150 countries, HiMedia is amongst the top three brands in the Bioscience Industry.
HiMedia Laboratories Private Limited is world renowned for manufacturing high quality culture media for microbiology. Additionally, we provide advanced media and products in the fields of Molecular Biology, Cell Biology, Plant Tissue Culture, Chemicals and Lab Aids/Equipment. As a Top Tier Global player, we are not only dedicated towards products but also striven towards introducing technologies such as Genomics Sequencing Services and Hydroponics.
HiMedia has managed to do this over decades as we have our own in-house bulk raw materials manufacturing plant. This enables us to deliver consistent quality products that conform to ISO 9001:2015 and ISO 13485:2012 and WHO: GMP.
HiMedia Labs. caters to one of the broadest Biosciences product categories: our premier established line of Microbiology products and newer promising products in Molecular Biology, Automated and Molecular Instruments, Cell Biology, Chemicals, and Premium Grade Lab Consumables, amongst others. The COVID-19 pandemic revolutionized not the clinical industry’s thought process regarding the significance of Molecular Diagnostics products.
The ‘Molecular Biology and Virology Division’ of HiMedia Laboratories Pvt. Ltd. Also called as HiGenoMB® is a One Stop Solution Provider churning out potential Research and Industry oriented Molecular biology products for the past glorious decade. About 2000 different products such as Nucleic Acid Extraction and Amplification (PCR) Kits, Cloning Reagents, Buffers & Chemicals for proteomics studies, Automated Molecular Instrumentation including RT PCR machines and PCR thermal cyclers and DNA/RNA Extraction platforms are being produced. The Proficient researchers in this department are spear heading the challenging field of Molecular Diagnostics to provide a complete solution for clinical diagnosis, agriculture, veterinary sciences, food industry, drug discovery and forensic medicine with the use of Real Time PCR or quantitative PCR kits and thermal cyclers. Our Molecular Biology Division-has established an in-house Advanced Sequencing and Bioinformatics facility which marks HiMedia’s entry into the Services space.
Our Cell Biology segment contributes with technologies which have brought in Serum free media for biopharma applications, Viral Vaccine Production Platform, Multicompendial grade chemicals, cultivated meat, and 3D bioprinting.
Moving from conventional to advanced automated methods like MALDI-TOF (Autof MS 1000) has been our newest endeavour for Microbiology.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.