Interview with Chris Moreland, Global Product Manager, Integrated Solutions, Promega, conducted by James Ives.
Why do scientists need to amplify bacterial DNA for molecular identification and serotyping of food-borne pathogens?
Identifying microbes is incredibly important in many industries and is often seen as quality assurance to make sure that the food manufactured or the ingredients used do not contain food-borne bacteria. Many states and countries also require testing for specific species, for example, Salmonella, Listeria, and E. Coli.

Image credit: Promega
Traditionally, methods for identifying microbes have used differential media. These are solid-state media that require incubation, sometimes overnight, sometimes for up to 48 hours. This can cause critical delays in identifying the microbes that may be present in a food sample. Using molecular techniques, especially amplification-based methods such as endpoint PCR or qPCR, provides faster results that allow decision making within hours rather than days.
Serotyping using amplification-based techniques (qPCR, WGS) also provides the ability to differentiate subspecies of a particular pathogen. For example more than 2500 subtypes of Salmonella can be differentiated using molecular methods, providing much more information than traditional techniques.
How is the Promega Maxwell® RSC Pathogen Kit used to purify bacterial DNA? How is this different from other techniques?
The Maxwell RSC PureFood Pathogen Kit is used with our Maxwell RSC instruments. These automated nucleic acid extraction systems use cellulose magnetic particles to bind DNA in a crude sample lysate, and then, through a series of wash steps, remove any inhibitors or materials that are not needed before eluting purified DNA ready for downstream amplification.
Image credit: Promega
The Maxwell RSC instrument is a cartridge-based system. All the reagents needed for purification are supplied in easy-to-use cartridges. The customer simply adds the sample to the cartridge, places it in the instrument, and runs the protocol. The system automatically performs the extraction, which is a major differentiator from other techniques. Automation eliminates much of the variability due to human error by removing manual-wash steps and pipetting associated with spin columns and other methods.
The Maxwell RSC PureFood Pathogen extraction protocol complements amplification-based tests, such as qPCR or NGS, which require high-quality purified DNA.
What advantages does the Maxwell RSC Purefood Pathogen Kit give researchers? How does this pathogen kit work?
One of the advantages to this technology is that it allows researchers to lyse both Gram-negative and Gram-positive bacteria in a single step. Many other kits make use of enzymes to digest cell walls and help break open Gram-positive bacteria. The lysis buffer used in the Maxwell RSC Purefood Pathogen Kit chemically dissolves those bonds to release the DNA from both cell types using a single step.
The use of laboratory automation removes variability, eliminating the inconsistency associated with manual techniques. Consistency is assured in the extractions, as well as in replicates.
Most importantly, by automating DNA purification and ensuring consistency between runs, Maxwell systems free up scientists to do other things, such as prepping samples or performing downstream analyses, providing more efficiency in their workflows.
What benefits does the Maxwell Purefood Pathogen kit have over other pathogen kits available?
With a differential medium, an "enrichment step" is done, where suspect food material is placed inside a Stomacher to allow any bacteria present to grow. The sample is then placed on differential media in an incubator. This process takes two days to complete.
With Maxwell systems, the overnight incubation enrichment step is still performed, but the subsequent DNA extraction is automated and multiple runs can be completed in an eight-hour shift. The Maxwell RSC instrument can process 16 samples, and the Maxwell RSC 48 can process 48 samples, in about an hour.
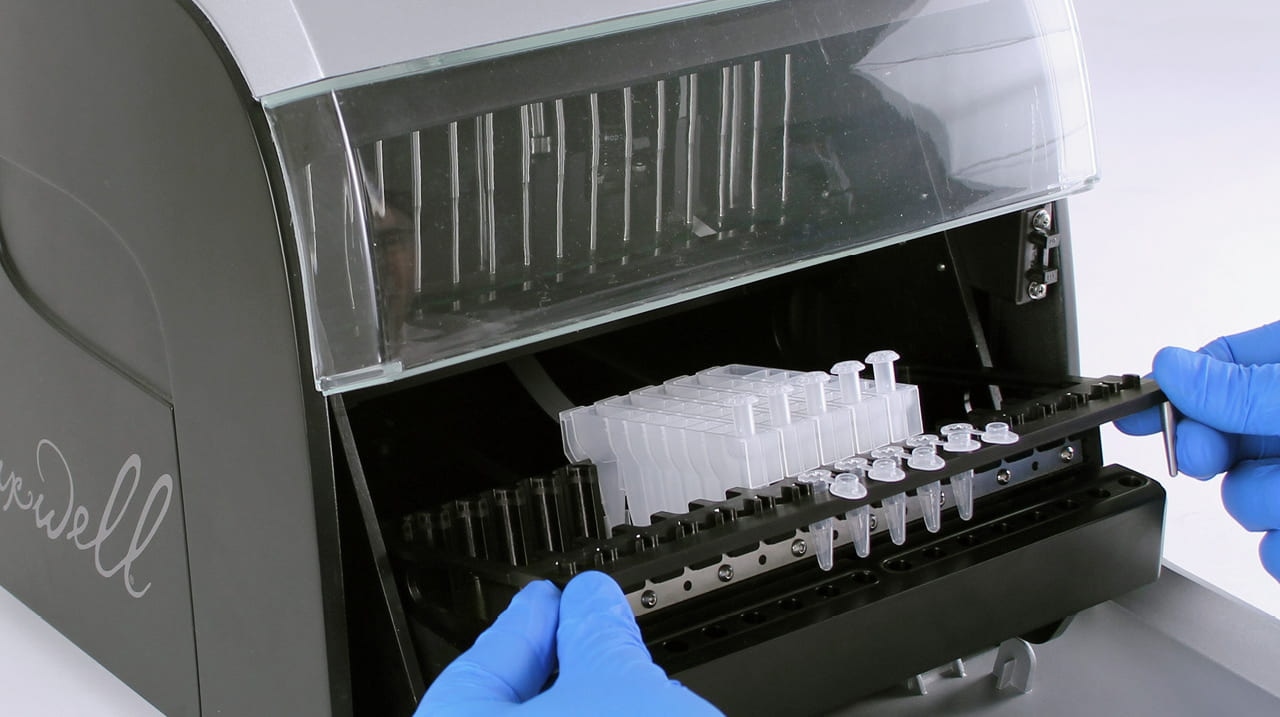
Image credit: Promega
Downstream qPCR analysis may take half an hour to 45 minutes. This automation allows researchers to process 16 to 48 samples, including downstream qPCR, in two or three hours instead of the two days required for manual methods.
It's key that laboratories move to more rapid methods of detection that provide actionable information in a faster time frame. If there are concerns about a contaminated piece of raw material or food that's been produced, the result can get back to either the manufacturer or to the lab quickly so that decisions can be made with minimal delay.
Can the Maxwell® RSC Purefood Pathogen kit be used with any other products to automate the process of pathogen extraction and identification?
We also offer a system called the MaxPrep, which can be used with the Maxwell RSC system for automated sample preparation, normalization and qPCR setup. MaxPrep is a liquid handler that has specific protocols to allow the researcher to do their qPCR setup.
MaxPrep is equipped with pre-set protocols, so the different steps do not need to be programmed in by the user. On the upstream side of the DNA extraction, MaxPrep can do the sample preparation and setup. The deck tray of the Maxwell RSC or RSC 48 is placed inside the MaxPrep, allowing integration of pre- and post-extraction steps, including dispensing of elution buffer and samples into the RSC cartridges.
Using the MaxPrep before and after extraction eliminates a lot of the pain points of manual -processing and is designed to automate the entire workflow.
Any DNA eluates that have been extracted are ready to use downstream in qPCR or endpoint PCR. The researcher can also detect or quantify the analytes using qPCR or absorbance, if needed.
In addition to expertise, extraction protocols and instrumentation, Promega also provides many of the buffers and enzymes needed for qPCR.
What assurances can Promega provide when using this pathogen kit and instrument?
All our reagents are tested to rigorous QA standards. All products are supplied with a Certificate of Analysis containing the details of all quality tests.
As a product manager I live for feedback, good and bad, not only to improve and develop our existing products, but also to fill gaps with products that we don't currently provide.
In the case of the Pathogen Kit, we worked closely with key opinion leaders in the industry early on, engaged with them to understand the challenges that they struggle with. They gave us some very direct feedback that we were able to incorporate into our product requirements during development. The final product is a culmination of that collaboration with the industry.
Can you give a brief overview of Promega, the work you do and the industries you serve?
Promega products are used by life scientists who are asking fundamental questions about biological processes as well as by scientists who are applying scientific knowledge to diagnose and treat diseases, detect microbial contamination in industrial and food manufacturing processes, discover new therapeutics, and use genetics and DNA testing for human identification.
Originally, founded in 1978 in Madison, Wisconsin, USA, Promega has branches in 16 countries and more than 50 global distributors serving 100 countries.
Where can readers find more information?
About Chris Moreland![]()
Chris received his MBA from Texas State University and his Bachelor of Science in Microbiology from Texas A&M University. In April 2013 he joined Promega as a Global Product Manager for nucleic acid purification and is responsible for managing the Maxwell® purification product line.