Spatial ecology or landscape ecology is a research area that investigates the formation of spatial patterns among bacterial communities. The magnitude of the 'grain' (the smallest unit being observed) and the 'extent' (the range of observations) are important determinants of the scale of examination. The scale is crucial when it comes to building theories about how such communities are organized.
How are such patterns formed? In the human mouth, at least, the answer includes factors such as the temperature, the moisture levels, the flow of saliva, the oxygen level, the pH, and the number of times abrasions or oral hygiene procedures occur. In addition to these macro-level factors, microbes themselves produce and utilize metabolic compounds, nutrients, and inhibitors, including antimicrobial molecules. They also physically prevent other microbes from occupying prime space, or their surfaces may offer good spots for other microbes to bind to. Such interactions lead to a diverse and functionally redundant community, which is more or less stable and metabolically active according to the levels of inter-microbial interaction.
To map the spatial orientation, other factors must be known, such as the distance between microbes as well as the distance between microbes and other host features such as the nearest host cell or the surface of a biofilm of which the microbe is a part. Imaging is used to obtain information on such patterns at individual cell level at scales of up to one millimeter.
The development of a technique called combinatorial labeling along with spectral imaging-fluorescence in situ hybridization (CLASI-FISH) has helped multiple microbial classes to be identified and localized at the same time by labeling any given type of microbe with multiple fluorophores. This helps visualize the spatial arrangement of a whole system of microbes forming microbial communities at the micron scale.
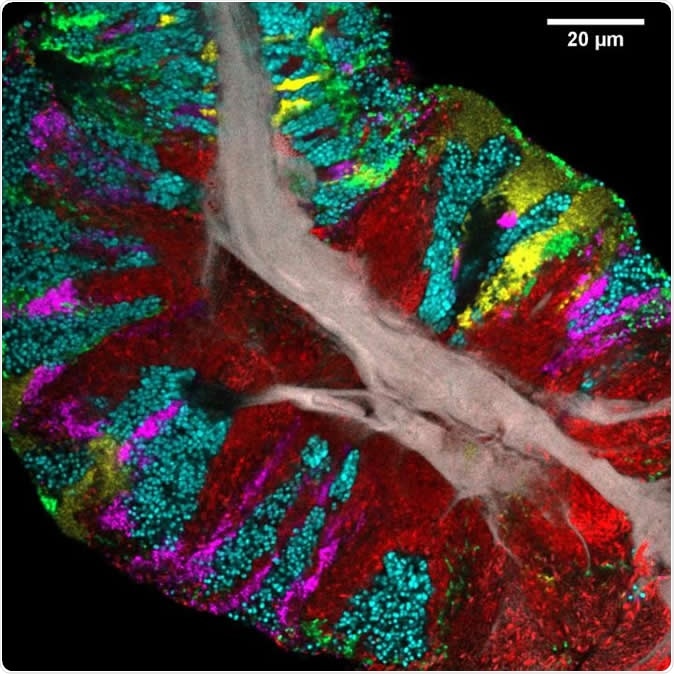
Bacterial biofilm scraped from the surface of the tongue and imaged using CLASI-FISH. Human epithelial tissue forms a central core (gray). Colors indicate different bacteria: Actinomyces (red) occupy a region close to the core; Streptococcus (green) is localized in an exterior crust and in stripes in the interior. Other taxa (Rothia, cyan; Neisseria, yellow; Veillonella, magenta) are present in clusters and stripes that suggest the growth of the community outward from the central core. Image Credit: Steven Wilbert and Gary Borisy, The Forsyth Institute
The study
The current study uses multi-spectral fluorescence imaging to establish its role in spatial ecology for microbial systems on the tongue. Here there are multiple densely clustered microbes in contact with the human epithelium and also with other oral habitats like the mucous membrane of the mouth and the teeth.
The investigators used a ridged plastic tongue scraper to collect a scraped specimen from back to front. The size and internal arrangement of these fragments of biofilm led them to conclude that they represented the spatial arrangement of the bacteria at various levels of the tongue dorsum faithfully over a scale of hundreds of micrometers. These levels include the tops of the filiform papillae covering the tongue, the valleys between them, and the fine spines projecting from them, all of which host bacteria of different types.
The researchers first identified the major bacterial types in the samples scraped from the tongues of 21 healthy volunteers by sequencing, and then analyzed each class to come up with a full view of the structure of the microbiome in sufficient detail to allow each of the key species to be assigned its own spot on the tongue.
Most of the microbial genes in the tongue community are formed of a limited number of oligotypes, according to the Human Microbiome Project (HMP). By linking each oligotype in the mouth to the bacterial classes in the expanded Human Oral Microbiome Database (eHOMD), the investigators identified 17 bacterial genera present in more than 80% of people and forming 0.5% of the microbes. Using sequencing data from the HMP, they found that 95% or more of the bacterial sequences came from a similar set of genera.
The researchers conclude that these genera are "likely to form both the spatial and the metabolic framework of the healthy TD microbiome."
Spatial organization
The researchers found three types of microbial arrangements: free bacteria, bacteria on squamous epithelial cells, and bacterial consortia, or structurally complex groups. The latter were bacterial biofilms made of several layers of microbes, with a distinct boundary and an epithelial core.
Bacterial composition analysis in each category and spatial location showed that consortia were more homogeneous than the other categories, with similar bacterial patterns across all samples. Free and epithelium-bound bacteria occurred singly or in small clusters. In contrast, each consortium displayed the same localized patch structure, each dominated by one bacterial type.
Each patch has a distinct boundary, is tens to hundreds of micrometers long, and has a core of human mucosal epithelial cells. The consortia live between the perimeter zone, which is exposed to saliva and oxygen, and the epithelial core.
At least one sample from every participant, and over 95% of sample images, showed the presence of 3 genera: Actinomyces, Rothia, and Streptococcus. Each had its own 'sweet spot' in the consortium, with Actinomyces forming large continuous domains near the core or stripes between patches of other bacteria. Rothia formed large patches near the perimeter as well as around a core of epithelial or bacterial cells. Within such a cortical layer, the Rothia was often broken up by streams or patches of other bacterial types. Streptococcus formed a thin external layer on the consortium as well as veins or patches inside it.
Other prominent bacteria types seen in samples from all individuals in the study included Veillonella, Gemella, Neisseriaceae, and phylum Saccharibacteria. Some of these bacterial classes may be to help convert nitrate in the saliva into nitrite and thus help regulate the levels of nitric oxide in the body. Less than a fifth of cells were not stained with any of the specific probes.
Next, the investigators looked at the different genera within the consortium, every species within one genus, and one species in particular which was thought to be the representative of that genus on the tongue. As expected from HMP data, they found, for instance, that Rothia was represented by R. mucilaginosa, Actinomyces by A. odontolyticus, and to a much lesser extent, A. graevenitizii, and Neisseria by N. flavescens. S. mitis, S. salivarius and S. parasanguinis were found on all consortia but in different places.
The formation of consortia
The scientists hypothesize that bacterial cells on the tongue dorsum push on each other as they multiply. Each class increases more rapidly in number in the area that is ideal for their growth, leading to unevenly shaped patches. This is the origin of the patch arrangement seen in the mature microbiome. In ill-health, the structure of the microbial community could vary.
"Our study is novel because no one before has been able to look at the biofilm on the tongue in a way that distinguishes all the different bacteria so that we can see how they arrange themselves," researcher Gary Borisy says. "Most of the previous work on bacterial communities used DNA sequencing-based approaches, but to get the DNA sequence, you have to first grind up the sample and extract the DNA, which destroys all the beautiful spatial structure that was there. Imaging with our CLASI-FISH technique lets us preserve the spatial structure and identify the bacteria at the same time."
In other words, this imaging method was able to identify most of the cells in each consortium, as well as their abundance and spatial arrangement in relation to the source of nutrition and the location of the substrate.
The micrometer-scale studies help differentiate microbial communities in relation to their biological arrangement. The use of species-level probes shows that many species are site specialists occurring at one site, like dental plaque, but not in the other, that is, the tongue dorsum.
The researchers conclude, "Although imaging is only one of several key technologies, it provides the unique benefit of showing us the target: the landscape and the structures that microbes build and that we need to explain and replicate in order to have achieved an understanding of microbial community."
Journal reference:
Wilbert et al., Spatial ecology of the human tongue dorsum microbiome, Cell Reports 30, 1–13 (2020), https://www.cell.com/cell-reports/fulltext/S2211-1247(20)30271-0