Even as the COVID-19 pandemic continues to spread to more and more people around the world, population-based measures to contain the outbreak are being reviewed. One important measure recommended by the World Health Organization (WHO) is enforcing quarantine for individuals or groups suspected of exposure to the virus until they are proved to be uninfected.
The length of the quarantine period is usually defined by the incubation period, from the time of exposure to the onset of symptoms. Each virus has a specific incubation period. If the quarantine period is too short, it could lead to unwanted exposure to the virus among healthy people. Too long a quarantine, on the other hand, could hurt the economy more than necessary as well as cause mental and emotional stress.
Predicting the Incubation Period
At present, there is no way to predict the incubation period of an unknown virus from its genome. The current study aims to fill the gap by providing a possible technique to arrive at a reliable and early estimate of this period and to set in motion a quarantine for possibly infected individuals.
The researchers looked at various genomic characteristics seen in single-stranded RNA (ssRNA) respiratory viruses to pick out those features that could be used to develop a model that predicts the incubation period. They then tested it out across a range of viruses and virus families to validate it. The model thus allows the viral incubation period to be accurately estimated.
The focus of the study was on this single group of viruses to eliminate as far as possible the confounding factors that could be the result of a different type of genome or different host tissue targets. The information on 14 viruses belonging to 4 different families was used.
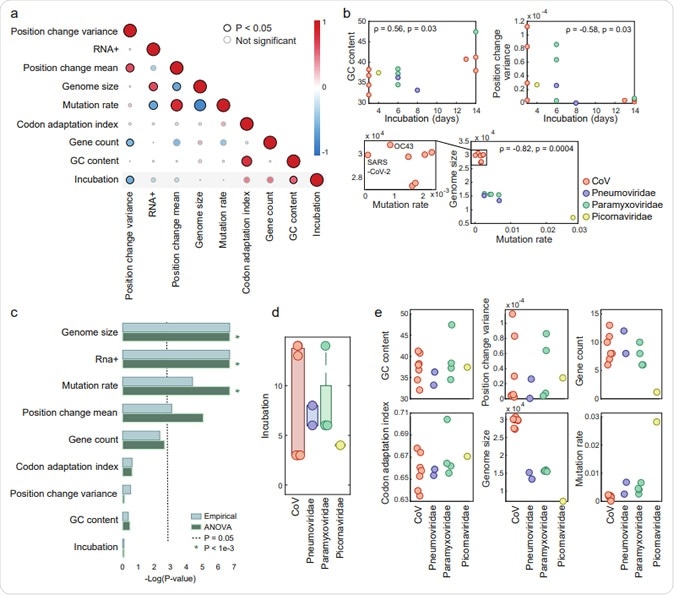
Genomic features of ssRNA viruses causing respiratory infections. (a) Pairwise correlation 105 matrix across all features. A description of feature construction is given in the Methods section. Each 106 circle indicates Spearman’s ρ between two features. The colors represent the rank-correlation 107 coefficients (red indicates positive correlation and blue indicates negative correlation), and the circle 108 sizes correspond to significance (p-value), where significant correlations (p-value <0.05) are circled in 109 black. (b) Scatter plots illustrating the relationships between features across four virus families. (c) 110 Estimation of the features association with the virus family, based on p-values (-log scaled) from two 111 tests applied (see Methods for details). The cutoff (p-value = 0.05) is indicated with a dashed line. Lower 112 values correspond to features that are not significantly associated with a virus family. (d) Boxplot and 113 overlaid dot plot of the incubation periods across viral families. (e) Dot plots of different features across virus families. The features shown in the upper panels are family-generic, and those in the bottom 115 panels are family-specific.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Using Family-Generic Features to Predict Incubation Periods
The analysis yielded the upper estimates for the incubation period for all the viruses. The researchers then selected eight features that could be responsible for the incubation period. These are based on the complete genome sequence and the way all strains of each type of virus align with the population genome.
Using pairwise analysis, the associations between these eight features, some already reported by earlier researchers, were shown to be valid. These include a lower mutation rate with higher genome length, but a higher codon adaptation index (CAI) with GC content.
The findings suggest a markedly lower rate of mutation of SARS-CoV-2 compared to the earlier SARS-CoV and other CoVs that cause human infection.
The next step was to choose only those features that could predict the incubation time, avoiding those that are chiefly due to the virus family. By avoiding the use of family-specific features and choosing family-generic features, they were able to train the predictive model.
The family generic features include:
- the GC content
- the differences in nucleotide number in each position following the alignment of the viral strains
- the number of genes coding for proteins
- the codon adaptation index
Using the dataset already analyzed to create training and testing sets, they trained one set on seven coronaviruses that infect humans, using the four family-generic features. Though this used only one virus family for training, the results were broadly applicable to the other families as well, with a mean absolute error of 1.6 days. This was closely correlated to the upper limit of the incubation period assigned in an independent data set.
The model predictions were also closely related to the ranks of the assigned incubation periods in the test set. The longest incubation period is known to be for measles, and this was so predicted as almost 10 days, with an upper limit of 14 days. Most reports of the incubation period of measles put it at 9-12 days. Similarly, the respiratory syncytial virus (RSV), which has the second-longest incubation period was also correctly defined as about nine days, with the assigned data putting it as eight days. The shortest incubation period was for rhinovirus, with 1.2 days, corresponding to the observed period of 1 day.
How Genome Factors Help Predict Incubation Period
The factors with the highest predictive power were the number of genes that code for proteins and the GC content, both being directly proportional to the incubation time. The mechanisms by which these associations operate are unclear. One could be that a more significant number of genes for translation means a longer replication cycle, while a higher GC content causing stable secondary structures to form within the viral RNA. This increases the energy barriers which are to be breached by the ribosome during translation time, increasing the latter. As translation cycles increase in length, so does the replication time, and thus the incubation time.
Another possible explanation is that when there are more genes, the host-virus interactions become complex, pushing up the incubation time. In fact, CoVs with high virulence also have long incubation periods and translate a higher number of accessory protein products, such as a protein with immunoglobulin-like domains. These are concerned with viral interactions with the host. In contrast, viruses with lower virulence have shorter incubation times.
Testing the Model on SARS-CoV-2
The predictive model was shown to be reliable by testing on training and test sets. Training with family-generic features allows the model to generalize to the test set, but when only family-specific features are used, this fails to occur.
The model was then tested to find out its potential for estimating the correct incubation period for the current SARS-CoV-2. Even though the training set used had only 2 viruses with incubation periods longer than 3 days, which could lead to underestimation of predicted incubation periods on the longer side, the model predicted that the SARS-CoV-2 would have an incubation period of about 9 days, which is in the upper range for incubation periods and covers the period within which most patients become symptomatic. Thus, this would have been a useful prediction for the COVID-19 pandemic in the early stages as well.
The Implications for Future Outbreaks
The study thus produced a predictive model based on genomic features that can predict virus incubation times for ssRNA viruses that cause human disease. The researchers identified four family-generic features of the genome that can be used to reliably and accurately predict the viral incubation period. These were used to build a model that can predict incubation periods and thus help to control future outbreaks like the current COVID-19 pandemic by providing an idea of the required quarantine period.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources