A startling new study by researchers at the Broad Institute of MIT and the University of British Columbia and published on the preprint server bioRxiv* in July 2020 casts doubt on the hypothesis that the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was transmitted from pangolins acting as the intermediate host. Instead, the authors suggest, the pangolins themselves were incidentally infected from other animals in captivity or from humans.
Since the current SARS-CoV-2 pandemic began, there have been several studies that showed the spike protein of the virus to have an almost identical genomic sequence with that of a Guangdong pangolin coronavirus (CoV). In fact, they share all five essential residues and have an overall 97% identical amino acids in the spike receptor-binding domain (RBD).
The importance of this discovery is that the viral spike protein is the binding site where the virus binds to the host cell. It is also a significant antigen and determines the host specificity of coronaviruses. The fact that pangolin CoV and the currently circulating virus had such high spike protein similarity seemed to indicate that this animal might have been the initial animal reservoir.
Bat CoV and SARS-CoV-2
Another closely related spike sequence among coronaviruses is found in the bat CoV, RaTG13, which is 90.1% similar in terms of amino acids. This virus has a 96% identical genome sequence, more even than the Guangdong pangolin strain, which has about 90% identity. The latter is still regarded as being probably the last intermediate host for the SARS-CoV-2 before its final leap across the species barrier to infect humans, because of the similarity of the spike RBD.
The Guangdong and Guangxi Viruses
The Guangdong pangolin CoV was isolated from a single batch of smuggled pangolins in the province of this name, in March 2019. It was first described in October 2019 by a scientist named Liu. Subsequently, several researchers have analyzed the genetic sequences of the CoV isolated from the same batch of animals.
However, another team of researchers described CoVs from another batch of pangolins from Guangxi province, but these had only 86% to 87% identity with the SARS-CoV-2.
The Study: Same Sequence, Different Papers
The current study aimed at examining the individual sequences from various studies, to find the one that represents the strain most broadly. This would be the best strain for further research use.
To their surprise, the researchers found that most of the pangolin sequences were simply repeated reconstructions or mapping of the same genetic material, based on the dataset supplied by Liu et al., in their metagenomic study in the journal Viruses.
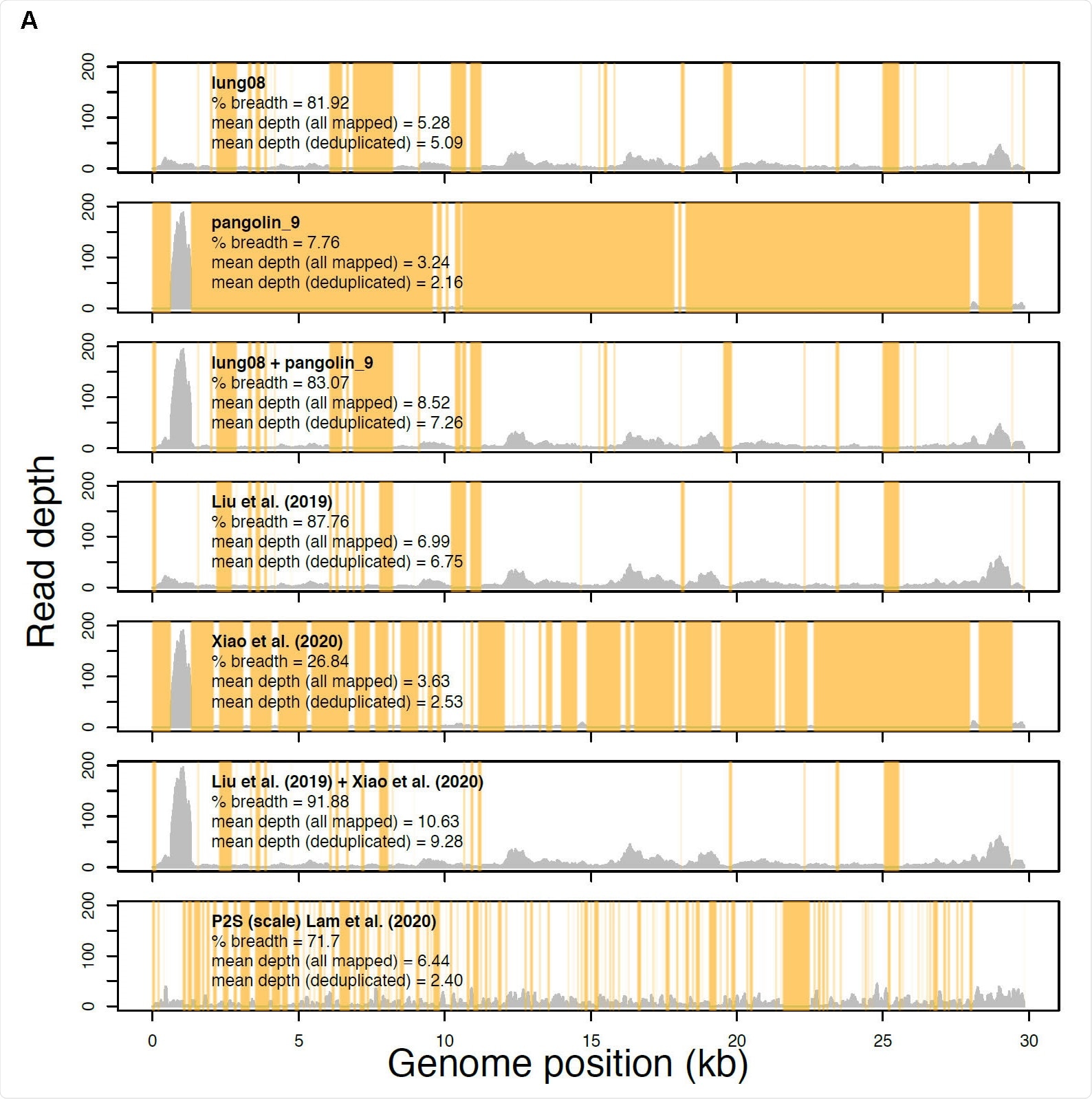
Read profiles of the metagenomic data sets from Liu et al. 2019 Viruses, Xiao et al. 2020 Nature, and Lam et al. 2020 Nature mapped to the Xiao et al. Guangdong pangolin CoV genome sequence GD_1 (EPI_ISL_410721). Samples lung08 (described in Liu et al. Viruses but re-introduced as M4 by Xiao et al. Nature) and pangolin_9 (sample M1, Xiao et al. Nature) each had the most sequence data of all samples analyzed in Liu et al. Viruses and Xiao et al. Nature, respectively. The “lung08 + pangolin_9” track shows their combined read coverage. The “Liu et al. (2019)” track indicates the read coverage pooled from all of the pangolin samples with mapped reads. The “Xiao et al. (2020)” track reveals the read coverage pooled from all samples unique to Xiao et al. Nature with mapped reads
Issues with Earlier Sequencing Publications
They begin with a paper authored by Xiao et al., in the journal Nature, showing that the sequences in this paper were the same as previously published by Liu et al., but without proper referencing. This makes it difficult to perceive their identity, especially since Xiao et al. also uses the term “total reads” in two different ways.
With two samples, the total reads refer to sequenced subgenomic fragments, but with the rest, this refers to the number of total reads, at two reads per fragment, which comes to double the library size. This had to be reduced to half before the pangolin samples from Liu et al. could be shown to match those used by Xiao et al.
Another issue is that one sample from Liu et al. (Viruses) and Xiao et al. (Nature) span the same sequences. Though the read depth of the metagenomic sequencing of the one sample from Liu et al. for the spike RBD was low, it supplied most of the sequence data. The sequences published by Xiao et al. are from samples that do not match those of Liu, though they are based on the same metagenomic analysis. They do not cover the spike RBD motif-containing amino acids that are critical for binding to the ACE2 receptor on the human host cell.
Again, Xiao et al. produced the full spike gene sequence using six pangolin samples infected with the virus. Still, the source data for the sequences of the spike RBD remain unpublished, except for the final sequence of GD_1.
The same paper by Xiao et al. shares another issue with that published by Liu et al. in the journal PLoS Pathogens, namely, the sequences used to fill the genome gaps by targeted PCR assays are not available. For this reason, the current researchers could not duplicate the genome sequence independently.
The method used by these groups is to assemble the genome by pooling sequences obtained from several different pangolin samples, arguing that all the samples contain the same type of CoV. Liu et al., in their second study, used 2/21 and 1/6 pangolins confiscated in March and June 2019, pooled to build one sequence.
These sequences were stated to be less abundant in the July 2019 sample compared with the March 2019 sequences, but have not been published. Thus, the bits of the Li et al. pangolin CoV genome that were taken from the July sequences remain unknown. The problem is, as the researchers explain, “Sequencing errors cannot be distinguished without access to the raw sequencing data, including the gap-filling sequences.”
And thus, say the researchers, it is important to understand one thing: “As expected, due to their reliance on the same dataset and, very likely, the same pangolin source, the Xiao et al. genome GD_1 (GISAID: EPI_ISL_410721) and the Liu et al. genome MP789 (the version that was updated on May 18, 2020) share 99.95% nucleotide identity.”
Non-Identity of Other Pangolin or Bat CoVs with SARS-CoV-2
In other words, the researchers say that there has been a single confirmed source of pangolin CoV with a spike RBD that is almost the same as that of the Guangdong pangolins. Further studies, such as those by Zhang et al. and Lam et al., also use the dataset of Liu et al. (Viruses). However, in the latter case, they acquired more pangolin samples from the Guangzhou Customs Technology Center. However, they obtained viral sequences from only one scale sample, and this was combined with that of Liu et al. to produce the pangolin CoV reference sequence.
Lam et al. sequenced Guangxi pangolin CoVs and found that the sequences are comparable in the spike RBD region, to both the Guangdong and the bat CoV RaTG13. However, only one of the five critical binding residues required for RBD-ACE2 engagement in SARS-CoV-2 is present in all three strains. The Guangdong pangolin CoV sequence has all five.
The authors say: “If there are batches of Guangdong pangolins other than the smuggled pangolins from March 2019 that have resulted in similar CoV sequences, particularly at the Spike RBD, we have not been able to locate such data based on the Liu et al. and Xiao et al. publications.”
Pangolins Could Be Incidental and Not Intermediate Hosts
As observed before, it needs to be clarified if the Guangdong and Guangxi strains are present in wild pangolins. While 14/17 of the infected pangolins in the first smuggled batch died within a month and a half, the virus was never again detected in a second batch of confiscated pangolins, or in a longitudinal study of 334 confiscated pangolins in Malaysia. This has led both the authors of the Malaysian study and the current study to put forward the hypothesis that the infected pangolins with identical viruses to SARS-CoV-2 were actually only an incidental host, having acquired the infection from humans or from some other infected animal.
If only smuggled pangolins contain the virus, the chances are that the infection came from another animal species and was acquired in captivity. This is especially likely since only one strain of the virus was recovered from each batch of smuggled pangolins.
Conclusion
The study concludes: “Although there is only a single source of pangolin CoVs that share a near-identical Spike RBD with SARS-CoV-2, and there is as yet no direct evidence of pangolins being an intermediate host of SARS-CoV-2, we would like to reinforce that pangolins and other trafficked animals should continue to be considered as carriers of infectious viruses with the potential to transmit into humans.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Sources:
- Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? Liu P, Jiang JZ, Wan XF, Hua Y, Li L, et al. (2020) Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)?. PLOS Pathogens 16(5): e1008421. https://doi.org/10.1371/journal.ppat.1008421
- Liu, P.; Chen, W.; Chen, J.-P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. https://www.mdpi.com/1999-4915/11/11/979
- Xiao, K., Zhai, J., Feng, Y. et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583, 286–289 (2020). https://doi.org/10.1038/s41586-020-2313-x
- Tao Zhang, Qunfu Wu, Zhigang Zhang, Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak, Current Biology, Volume 30, Issue 7, https://doi.org/10.1016/j.cub.2020.03.022.
- Lam, T.T., Jia, N., Zhang, Y. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020). https://doi.org/10.1038/s41586-020-2169-0
Journal reference: