As the COVID-19 pandemic continues to spread in many regions of the world, it is becoming clear that better indicators are required to differentiate severe disease from mild to optimize the use of resources in the treatment of this condition. Two University of New Mexico researchers postulate certain specific biomarkers' relevance in predicting the potential for progression to severe COVID-19 in a paper published on the preprint server bioRxiv*.
The clinical features of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection vary since it can cause asymptomatic, mild, moderate, or severe disease, including multi-organ injury and death. A poor prognosis is more common in those over 65 years, individuals with underlying chronic illness (diabetes, cardiovascular disease, hypertension, obesity, and chronic obstructive lung disease) and elevated levels of certain inflammatory molecules like IL-6, CRP, and D-dimer.
Severe COVID-19 is known to be linked to the cytokine storm syndrome, in which multiple inflammatory molecules are released in an unregulated manner, leading to acute respiratory distress syndrome (ARDS), the most common cause of death in these patients. The occurrence of pre-existing cross-reactive T cells and antibodies in healthy individuals does not explain how older people are at increased risk since they have a higher probability of pre-existing immunity. Nor does this explain the known male predilection for more severe disease.
Lower Th17, Increased IgA-Secreting B Cells
The current study takes a new look at earlier single-cell RNA sequencing data, comparing findings from the bronchoalveolar lavage fluid (BALF) of patients with severe disease and those with mild disease. They found that immune cells, inflammatory pathways, and enzymes were dysregulated in the former.
There was a fall in the frequency of Th17 cells in severe disease but an increase in IgA-secreting B cells. This phenotype of SARS-CoV-2 infection has been associated with dysregulation of myeloid and CD8+ T cell lines, with a more significant number of macrophages and neutrophils but fewer CD8+ T cell subsets.
Earlier studies indicate that the lung tissue shows a hyperinflammatory profile relative to either healthy controls or patients with community-acquired pneumonia. Like the earlier SARS virus, this one strongly activates the expression of a number of IFN-stimulated genes (ISGs), with elevated cytokines like IL1B, IL6, and TNF.
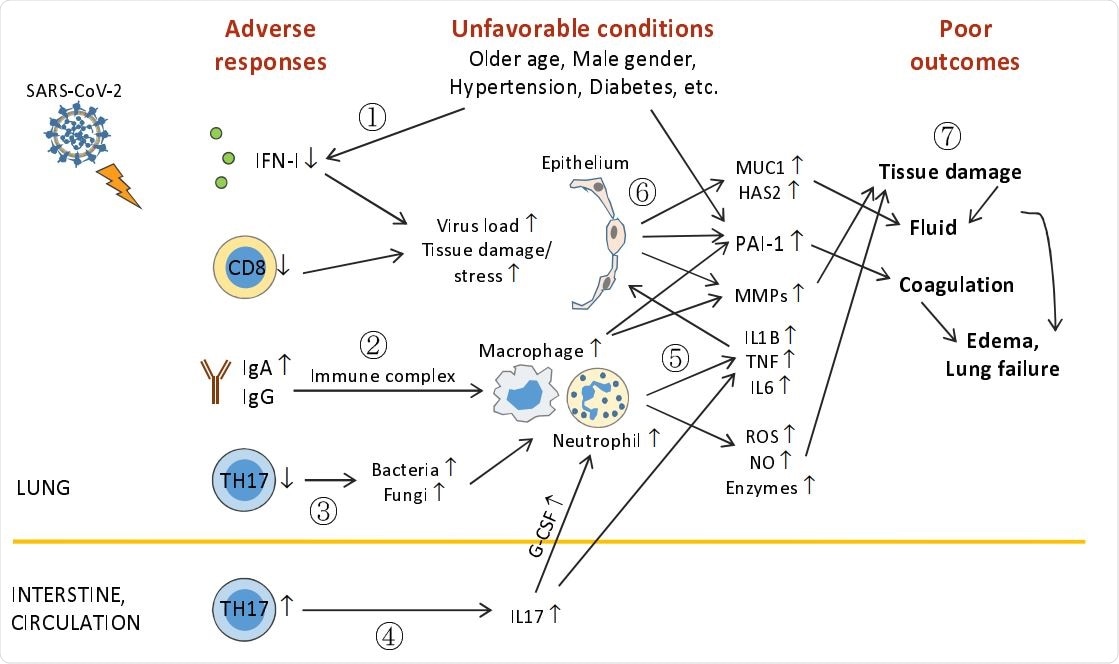
Outline of unfavorable conditions and deleterious pulmonary responses. (1) Older age, male gender, underlying conditions (such as hypertension, diabetes, etc.), and unknown factors (including genetic background) impair anti-viral immunity (including IFN-I deficiency and decreased CD8+ T and TH17 cells), leading to higher virus loads and tissue damage/stress. (2) Elevation of humoral responses results in massive immune complexes that activates macrophages and neutrophils. (3) Decreased TH17 cell responses cause overgrowth of commensal bacteria and fungi, which further activate macrophages and neutrophils. (4) TH17 hyper-activation and/or expansion in the intestine (?) cause high levels of serum IL17, which induces G-CSF expression and in turn, promotes neutrophilia. (5) Hyper-activated macrophages and neutrophils release immense amounts of proinflammatory cytokines, leading to cytokine release syndrome and subsequent ARDS, and tissue destructive products, such as ROS, NO, MMPs, and other enzymes. (6) During ARDS, proinflammatory cytokines act on epithelial cells and induce MMPs, mucins, hyaluronic acids, antimicrobial peptides, and PAI-1 (unfavorable conditions also elevate PAI-1 expression). (7) ROS, NO, MMPs, and other enzymes cause epithelial and endothelial leakage, leading to tissue fluid/plasma accumulation in alveolar spaces. Mucins, hyaluronic acids, and antimicrobial peptides concentrate alveolar fluids and thicken mucosal lining, resulting in drowning edema and even lung failure. Heightened PAI-1 facilitates coagulation and strengthens edema formation (and thrombosis). ARDS also has systemic consequences causing multi-organ damage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Reduced Th17 Expression
Another study found comparable levels of Th1, Th2, and T regulatory cells in mild vs. severe disease, in the CD4+ T cell compartment. The significant differences include a reduction in Th17 cells and γδT cells, as well as lower levels of the Th17 cytokines expressed by the latter.
Th17 cells are considered to be important in tissue pathology, but they are also crucial to antiviral immune responses since they trigger Th1 responses. This type of immunity involves cytotoxic T cell and B cell responses, typically seen in bacterial infections.
Severe COVID-19 is characterized by poor Th17 responses, which may mean these cells protect the body against infection. This could indicate the potential for the early use of antibiotics in such patients.
The gut also shows significant Th17 responses, and the SARS-CoV-2 receptors, ACE2 and TMPRSS2, are expressed in intestinal cells. Overall, therefore, the researchers comment, “The systemic role of TH17 cells in the disease progress, especially the development of ARDS, need further define.”
The cytokine IL22 was seen in most severe, but none of the mild disease cases and its role needs to be examined further. Another change that was seen in mild vs. severe disease was the higher number of IgA1- and IgG1-secreting B cells in the latter. This flood of antibodies in severe disease may indicate the potential for large-scale immune complex formation and deposition and resulting in pathologic inflammation. This hike in tissue permeability is characteristic of autoimmune conditions like systemic lupus erythematosus (SLE).
Other researchers have noted that the higher the titer of specific anti-SARS-CoV-2 antibodies, the more severe the disease.
IFN-I Response in Severe COVID-19
The IFN-I pathway is an essential antiviral mechanism. In severe COVID-19, IFNA1 is found to be decreased in the macrophages, while the other interferon pathways, IFNA2 and IFNB1, are expressed more highly. IFNA2 levels were also higher in women.
The Toll-like receptors TLR7 and TLR8 are also downregulated in macrophages and in the epithelium. These are the receptors that sense the presence of RNA.
IFN-I is also reduced in diabetes and hypertension. Work with viruses such as the flu virus or the West Nile virus has shown that in older people, the dendritic cells secrete less IFN-I, which, in turn, reduces the secretion of IFNγ in CD4 T cells and IFNγ, perforin and granzyme in CD8 T cells.
The researchers say, “These results implicate an essential role of IFN-I pathway in the disease susceptibility.”
Higher MMP in Severe COVID-19
With severe COVID-19, the MMPs in epithelial tissue are found to be raised from double to almost eight-fold the levels in mild disease. High MMP activity is a finding associated with ARDS, chronic obstructive lung disease, tuberculosis, and interstitial pulmonary fibrosis, all of which destroy the lung eventually due to the way they break down the matrix molecules of the extracellular space in the interstitial and tight junction spaces.
These enzymes are inhibited by tissue inhibitors of metalloproteinases (TIMPs), which are found to be unaltered in the epithelium. Macrophages in severe COVID-19, however, had a higher percentage of TIMP1 but lower TIMP2 cells. Overall, the upregulation of MMPs, macrophages and neutrophil oxidizing radicals could be related to the serious lung destruction seen in severe COVID-19.
Increased MUC1, HAS2, and PAI-1 in Severe COVID-19
High Mucin-1 levels are found in several lung diseases, such as viral infections, asthma, and chronic obstructive pulmonary disease (COPD). However, the difference between mild and severe COVID-19 was not significant in epithelial cells. Hyaluronic acid was secreted at higher levels in severe disease, with HAS2 being upregulated in the epithelium. Other HAS genes were not affected.
The expression of tPA and PAI-1 was also elevated in severe COVID-19, with the latter being high in both epithelial cells and macrophages. The ratio of tPA to PAI-1 cells went down in severe disease, indicating loss of regulatory control of coagulation. Indeed, coagulation failure is a cause of worse outcomes in severe COVID-19.
PAI-1 is upregulated by many inflammatory molecules such as IL1, IL6, TNF, and TGFβ. Again, it is elevated in response to hormones, such as insulin, glucocorticoids, and epinephrine. It goes up with age, and this is linked to cardiovascular and metabolic disease in advancing age. PAI-1 is also elevated in men relative to women. Thus, PAI-1 elevations may signal the potential for severe COVID-19, and all the more if the above conditions are met.
The researchers sum up: “Augmented expression of MUC1, HAS2 and PAI-1 is associated with more severe disease and may contribute to the drowning edema and the coagulation disorder in severe COVID-19.”
Implications
As a result of these analyses, the study indicates that pulmonary antiviral defenses are dysregulated following SARS-CoV-2 infection. There is a fall in Th17 cells, CD8 T cells, and in IFN-1. Of these, the decrease in IFN-I is associated with impaired TLR7 and TLR8 expression.
At the same time, they identified other predictors such as IgA, MMPs, and MUC1 and found them to correlate with the severity of COVID-19 and other infectious diseases. The markers IFN-I, with TLR7 and TLR8, and PAI-1, may help predict the risk of severe COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources