The COVID-19 pandemic sweeping through the globe has led to enormous efforts to understand the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes the disease. One effort toward treatment has been the repurposing of autophagy-modulating drugs like chloroquine and hydroxychloroquine. However, trials show these drugs have no significant benefits for treating the disease.
Autophagy is a cellular mechanism that removes damaged or unnecessary cell components, allowing the body to get rid of these cells and, in addition, acting against infecting pathogens. The process is highly regulated and is affected by different factors like oxidative stress, deficiency or excess of nutrients, and viral infection.
The serine/threonine unc-51-like kinase (ULK1) plays a critical role in autophagy and is believed to be involved in many diseases like cancer, neurodegeneration, and inflammatory disorders. ULK1 has an N-terminal kinase domain and C-terminal early autophagy targeting domain that helps ULK1 interact with its various substrates.
Positive-sense RNA viruses like betacoronovirus use autophagy to utilize cellular double-membrane vesicles as surfaces for RNA synthesis, but how they subvert autophagy components for their use is not well understood.
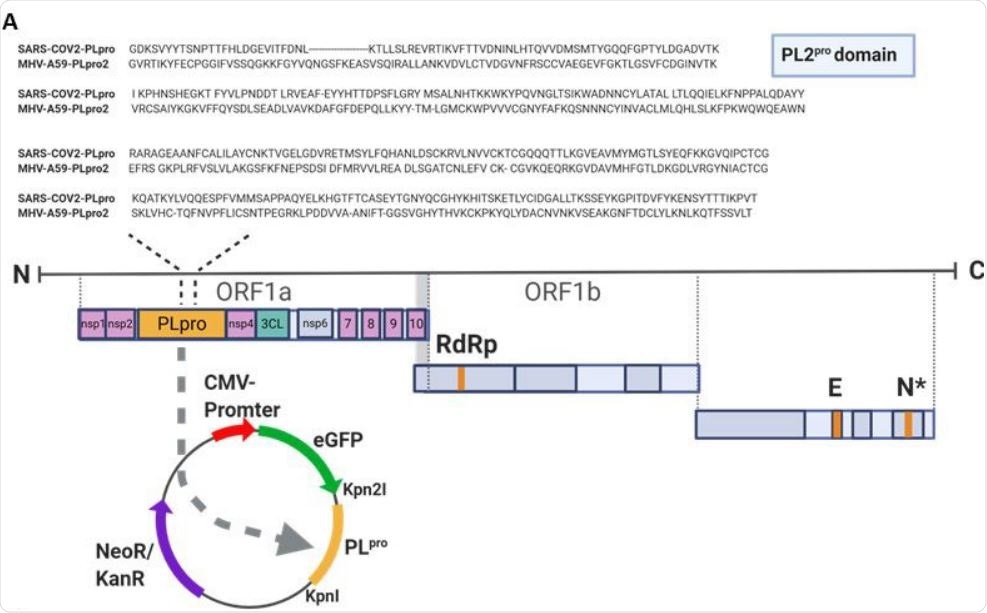
PLpro of SARS-CoV-2 cleaves ULK1 (A) Sequence alignment of the PL protease domain of SARS-CoV-2 and PL protease domain 2 of MHV-A59 is shown. The SARS-CoV2-PLpro was cloned into EGFP-C1 vector.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Papain-like protease targets ULK1
Researchers from the University of British Columbia have discovered that the betacoronavirus-encoded papain-like protease disrupts autophagy, partly by targeting ULK1. Their work is reported on the preprint server bioRxiv* in October 2020.
To understand the regulation and function of ULK1 during betacoronavirus infection, the researchers used mouse hepatitis virus A59 as a model and an infected murine fibroblast cell line 17C11.
They observed loss of ULK1 starting about 12 hours after infection using anti-ULK1 antibodies. A previous study also found reduced ULK1 levels during porcine betacoronavirus infection. Using polyclonal antibodies, they observed a potential cleavage fragment that was not detected using monoclonal antibodies.
This prompted the scientists to look into whether betacoronavirus proteases might be responsible. Of the two proteases found in the viruses, the team made a construct expressing the papain-like cysteine protease of SARS-CoV-2, which has 63% similarity to that of the mouse virus.
To determine if the papain protease targets ULK1, they used HEK293T cells. Again, they found reduced ULK1 levels and a smaller molecular weight cleaved fragment.
The team also tested to see if the other protease in betacoronaviruses, a chymotrypsin-like cysteine protease, was responsible for the reduction in ULK1 levels or cleavage using an in vitro assay. They found this protease was not involved in ULK1 cleavage, suggesting the effect of only the papain protease.
Next, the researchers determined the location on ULK1, where the papain protease acts. Using previous studies, they identified two potential cleavage sites. The team mutated the potential cleavage sites, and transfected wild-type or mutant ULK1 along with control or the papain protease construct into HEK293T cells.
They found cleavage in the wild-type and G531 mutant ULK1, but not in the G499 mutant. This suggests the protease cleaves ULK1 after G499, separating the N-terminal kinase domain from the C-terminal domain that binds to substrates.
Furthermore, the scientists found that the cleavage results in reduced co-immunoprecipitation of ULK1 with ATG13, a molecule that binds to ULK1 and a protein in the autophagy-initiating ULK1 complex, suggesting cleavage of ULK1 results in reduced binding. The ULK1 complex is required for starvation-induced autophagy.
The cleavage resulted in reduced autophagic degradation, and they found that cells expressing the papain protease did not initiate autophagy.
SARS-CoV-2 disrupts cellular autophagy
When a ULK1 inhibitor was added to cells infected with the mouse virus, the team found that viral RNA replication was reduced significantly. Furthermore, a non-cleavable ULK1 mutant suppressed viral replication compared to the wild-type ULK1. This indicates betacoronaviruses need ULK1 before cleavage for replication.
Previous studies on SARS-CoV reported deubiquitinase activity by recognizing the LXGG motif. The cleavage site of the ULK1 in SARS-CoV-2 after glycine is also similar. Targeting ULK1 and other host proteins with LXGG could be another new mechanism that betacoronaviruses use to disrupt cellular autophagy.
ULK1 levels started decreasing later in the mouse virus infection, coinciding with the observation of cleavage fragments. This suggests ULK1 function, although necessary for viral replication in the initial stages of infection, might be dispensable in later stages of viral infection, or cleavage may have new functions that serve the virus. Non-cleavage of ULK1 showing low virus replication suggests continued ULK1 activity throughout the infection may be bad for the virus.
Thus, the study provides new mechanisms by which the SARS-CoV-2 virus uses papain protease to disrupt cellular autophagy.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources