The devastating impact of the coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to significant efforts to curb its transmission. When RNA viruses, like the SARS-CoV-2, replicate there could be versions of the virus that have large deletions in the viral genome, making the virus defective. These defective genomes can still replicate in the presence of the full-length virus. Because they are shorter, they can replicate faster than the original virus, and compete with the full-length virus for replication, disrupting the virus's growth and spread.
.jpg)
3D rendering of SARS-CoV-2 microbe with RNA molecule inside. Image Credit: Vchal / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
These defective interfering (DI) genomes are common in coronaviruses, but DI has not yet been reported for SARS-CoV-2. DI particles are generally believed to be a result of inefficient replication or having regulatory functions. Researchers from Pennsylvania State University, USA, have developed a synthetic DI construct for SARS-CoV-2 and reported their findings in a new study published on the bioRxiv* preprint server.
A synthetic defective interfering genome
They prepared synthetic DI genomes from parts of the SARS-CoV-2 genome and tested if the DI genomes could replicate in cells infected with both the synthetic genome and the wild type (WT) virus.
They made their construct using three portions of the virus genome: the 5' UTR and the 5' part of the non-structural protein 1 (nsp1) in ORF1a, a part of nsp15 with the packaging signal, and the sequence of the 3' part of the N sequence, ORF10 and 3'UTR. The synthetic DI genome was 2,882 nucleotides (nt) long, about 9.6% of the WT virus's genome length. In addition, the authors also prepared a second shorter synthetic genome about 800 nt long without the packaging portion.
The team prepared the two genomes as DNA and inserted them into plasmids, transcribing them in vitro to make the RNAs. The prepared RNAs were inserted into Vero-E6 cells infected with SARS-CoV-2.
Because the synthetic DI genomes degrade quickly and do not replicate well, the authors were not able to study how they replicated. However, DI genome with the three portions reduced SARS-CoV-2 replication by about half, 24 hours after transfection. The shorter DI genome did not have any effect on SARS-CoV-2 growth.
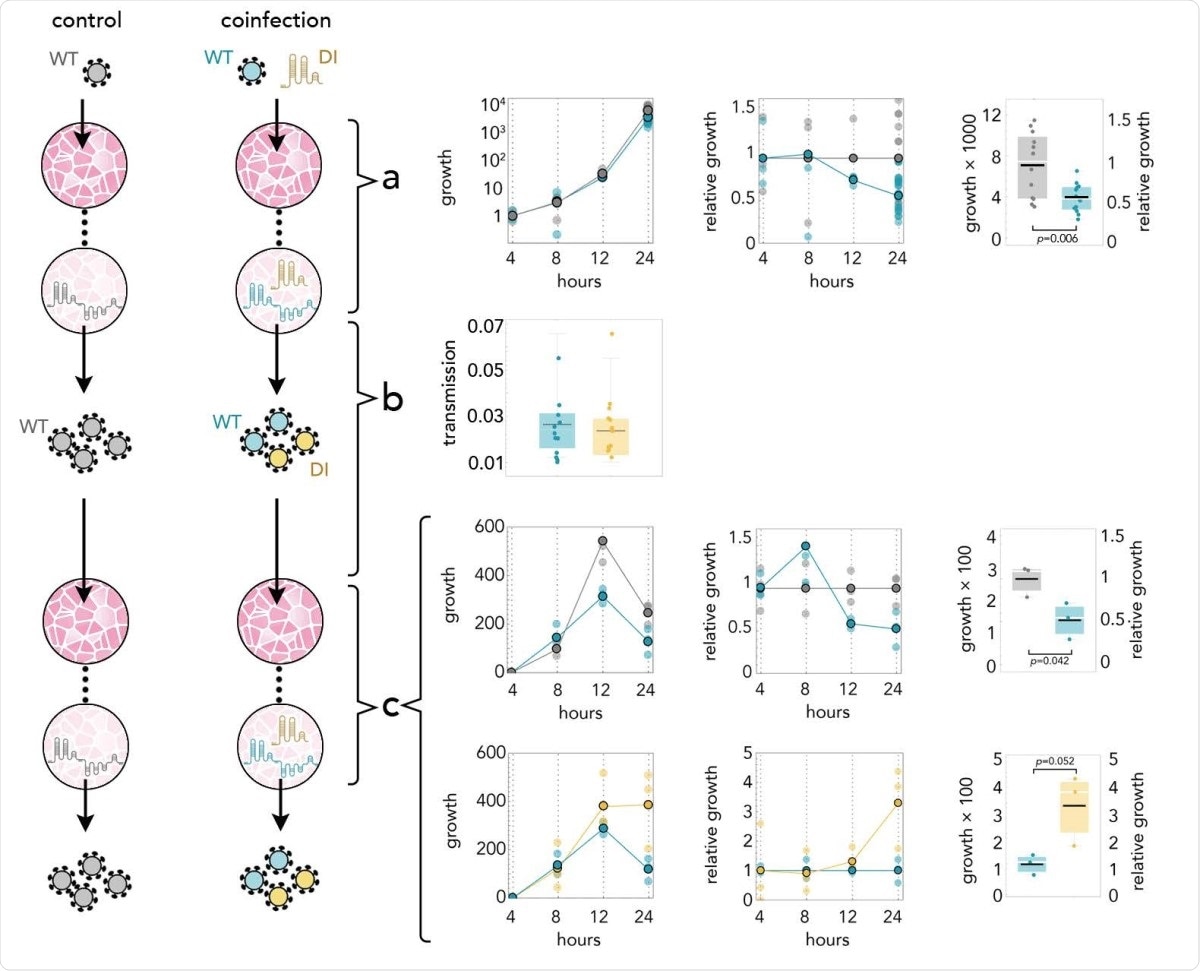
DI reduces the amount of SARS-CoV-2 by half; it replicates 3 times faster; and it is transmitted with the same efficiency. Yellow: DI in coinfections; blue: WT in coinfections; gray: WT in infections without DI. a: Growth rates (absolute amount relative to the amount at 4 hours) of WT in controls and in coinfections; growth relative to controls at the same time point; and detail at 24 hours. b: 24 hours after infection the supernatant was used to infect new cells. The transmission efficiency is the amount measured by qRT-PCR immediately before passaging divided by the average amount measured almost immediately (4 hours) after passaging. c: Growth rates (absolute amount relative to the amount at 4 hours) of WT in controls and in coinfections; growth relative to controls at the same time point; and detail at 24 hours. Growth rates (absolute amount relative to the amount at 4 hours) of WT and DI in coinfections; growth relative to that of WT in coinfections at the same time point; and detail at 24 hours.
A day after transfection, the team infected new cells using the supernatant. Both the WT virus and the DI genome were seen starting four hours after infection, but they did not see the DI genome without the packaging portion. There was no difference in the DI genome's transmission rate and the WT virus, suggesting the shorter DI genome is as infectious as the WT virus.
In the samples infected both with the DI genome and the regular virus, the number of regular virions decreased by half in 24 hours. The DI genome increased about three times as fast as the WT virus. The results suggest that even a small amount of DI genome can strongly affect replication of the WT virus.
Defective genome competes with normal virus during growth
The authors also modeled the dynamics of the system to understand the intra-cell competition between the DI genome and normal virus. Similar to predictions from previous models, DI genome and WT virus coexist if the replication advantage of the DI genome is below a critical threshold. Above that threshold, the DI genomes will kill off all WT viruses. This threshold depends on the number of genomes within the range of the viral protein produced by the regular virus.
Thus, the model suggests the DI genome will increase over time and kill off the WT virus genome. However, since the authors measured only five passages, where they did see an increase in the DI to WT genome ratio, they write they were not able to verify if this early reduction would lead to the extinction of the WT virus.
"The interference with the WT virus is the most remarkable effect of our DI construct," write the authors. DI particles do not serve any purpose for the WT virus; rather, they exist as parasites to the normal virus. Because they can replicate faster than the full-length virus when co-infected with the virus, they are suitable as potential antivirals.
As the replication goes on, the DI genome continues increasing in frequency, and the process becomes more effective, ultimately leading to the death of both the normal virus and the DI genome. Although DI constructs have potential as antivirals, they have not been explored much. HIV and the influenza viruses are not ideal candidates for this approach as they have short genomes and complex life cycles. However, coronaviruses are ideal for this approach as they have a long single-stranded RNA genome and simple life cycles and could be explored further for SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources