Researchers studying the effect of convalescent sera and monoclonal antibodies from recovered patients found the UK and South African SARS-CoV-2 variants are more resistant to neutralization compared to the wild-type virus.
As the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to spread across the globe, it has evolved and undergone several mutations. The new virus strains may have different transmissibility and infectivity which may affect how neutralizing antibodies work against them.
There have been reports of a decrease in antibody response to the virus, and some reinfection cases have been reported. Antibodies to the SARS-CoV-2 spike protein are the major proportion of neutralizing antibodies. The virus variants originating recently in the United Kingdom, B.1.1.7, and South Africa, B. 1.351, have several mutations on the spike protein. This is concerning with regards to the effect of neutralizing antibodies.
Researchers from Chongqing Medical University in China studied how effective convalescent sera and monoclonal antibodies are to these variants. They published their results in a research paper posted on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Mutant viruses resistant to some antibodies
The team used previously obtained blood samples from 20 COVID-19 patients in February 2020 and then again in October 2020, after their recovery, to collect eight receptor-binding domain (RBD) specific monoclonal antibodies. They expressed the wild type and the UK and South African mutant strains in a lentiviral pseudotype system.
The authors assessed the infectivity of the variant strains using luciferase assay. They found the two variants had entry efficiencies 3–4 times that of the wild type virus. This suggests the variants have higher infectivity than that of the wild-type virus.
The sera collected from patients about 25 days after symptom onset were less effective in neutralizing the mutants than the wild-type virus. Neutralizing antibody titers were 825 for the wild-type and 343 and 148 for the UK and South African variants, respectively.
When tested against sera collected about eight months after symptom onset, 85% of the samples showed inhibitory dose (ID50) greater than 40 to the wild-type virus. About 40% and 90% of the samples had ID50 titers below the UK and South African mutant threshold, respectively. Thus, some convalescent sera are not able to neutralize the variants.
Next, the authors tested how effective monoclonal antibodies collected from the convalescent sera were in neutralizing the different virus strains. Of the eight types of antibodies collected, all had strong neutralizing activity on the wild-type virus. While the variants had almost no effect on two antibodies, the other six had diminished neutralizing activity against the variants.
Of these six, three and five were less effective by 3-folds or more against the UK and South African strains, respectively. Two antibodies had no effect on the South African variant, and it reduced the effect of the most potent antibody 26 folds compared to that of the wild-type virus.
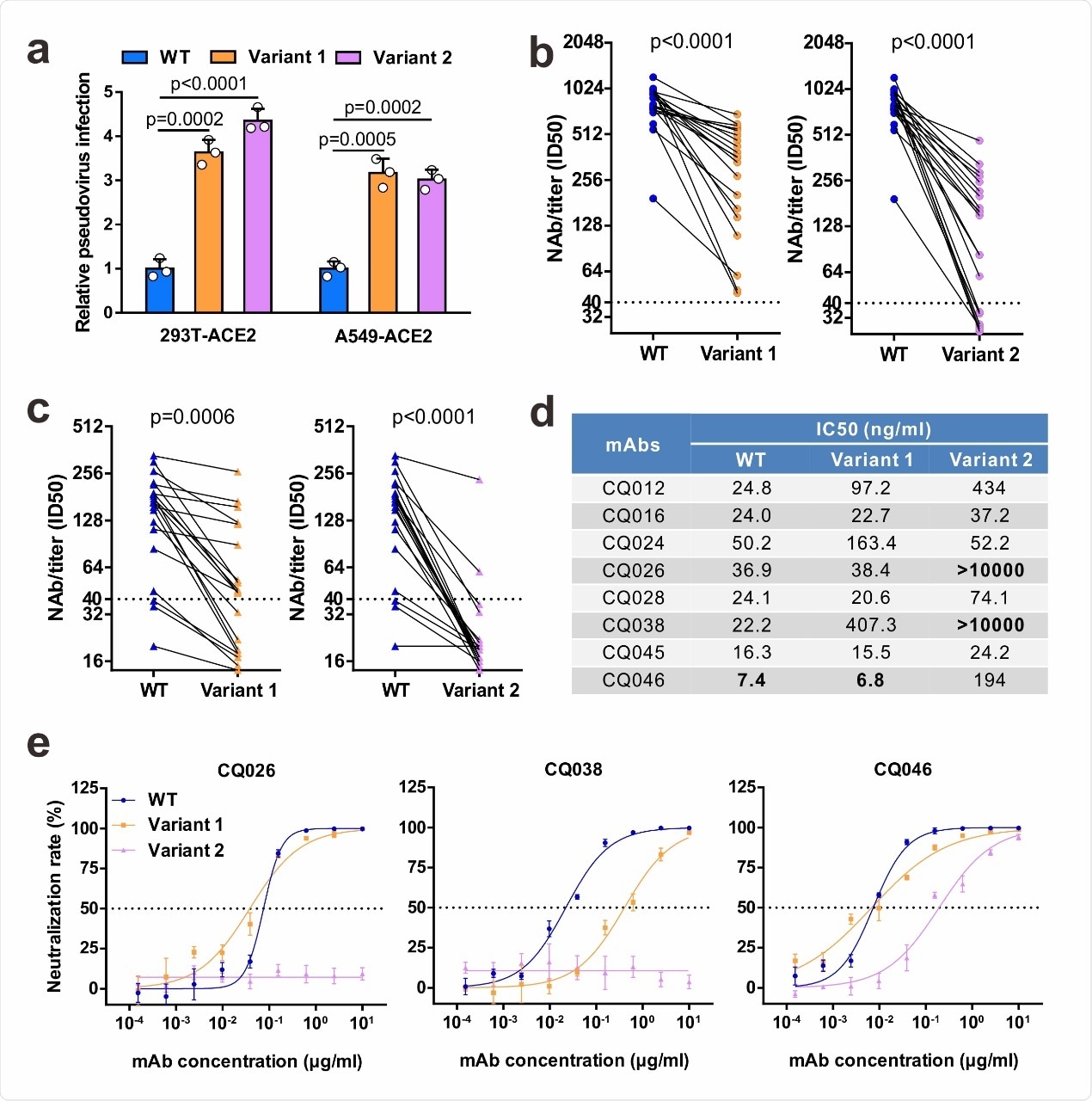
Neutralizing activities of convalescent sera and monoclonal antibodies against SARS-CoV-2 variants. a Infectivity of WT and variant pseudovirus conducted in 293T-ACE2 and A549-ACE2 cells. Cells were inoculated with equivalent doses of each pseudotyped virus. WT, wild-type Spike (GenBank: QHD43416) pseudotyped virus; Variant 1, N501Y.V1 mutant Spike pseudotyped virus (containing H60/V70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H); Variant 2, N501Y.V2 mutant Spike pseudotyped virus (containing K417N, E484K, N501Y, D614G). b-c Pseudovirus-based neutralizing assays were performed to detect neutralizing antibody (NAb) titers against SARS-CoV-2. The thresholds of detection were 1:40 of ID50. Twenty sera (indicated by circles) were drawn 5 to 33 days post-symptom onset (b); 20 sera (indicated by triangles) were drawn ~ 8 months post-symptom onset (c). d-e The half-maximal inhibitory concentrations (IC50) for tested monoclonal antibodies (mAbs) against pseudoviruses (d) and representative neutralization curves (e). Statistical significance was determined by One-way ANOVA
Implications of new variants on antibody neutralization
These results show that both the variants increase viral infectivity compared to the wild-type strain. Both the variants have the mutations D614G and N501Y in the spike protein, which have been associated with an increase in viral fitness and transmissibility.
The inability of the convalescent sera and monoclonal antibodies derived from patients who were infected during the first wave of infections in China suggests these variants may have evolved to escape neutralization by antibodies. However, it is yet unknown if these patients will be at higher risk of reinfection by these variants.
“Given the evolving nature of the SARS-CoV-2 RNA genome, antibody therapeutics and vaccine development require further considerations to accommodate mutations in Spike that may affect the antigenicity of the virus,” write the authors. Further studies with a larger sample size and using the complete SARS-CoV-2 virus and mutant strains, rather than the pseudoviruses, will help further our understanding.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hu, J. et al. (2021). Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. bioRxiv, https://doi.org/10.1101/2021.01.22.427749, https://www.biorxiv.org/content/10.1101/2021.01.22.427749v1
- Peer reviewed and published scientific report.
Hu, Jie, Pai Peng, Kai Wang, Liang Fang, Fei-yang Luo, Ai-shun Jin, Bei-zhong Liu, Ni Tang, and Ai-long Huang. 2021. “Emerging SARS-CoV-2 Variants Reduce Neutralization Sensitivity to Convalescent Sera and Monoclonal Antibodies.” Cellular & Molecular Immunology 18 (4): 1061–63. https://doi.org/10.1038/s41423-021-00648-1. https://www.nature.com/articles/s41423-021-00648-1.