The coronavirus disease 2019 (COVID-19) pandemic has resulted in over two million deaths worldwide, out of over a hundred million reported cases to date. The virus that causes it, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), binds to the host cell by its receptor, the angiotensin-converting enzyme 2 (ACE2). It is reported that the UK and South African strains may have higher transmission capabilities due to amino acid substitutions on the SARS-CoV-2 Spike protein. Although disease pathogenesis remains unclear, in addition to the host response mediated by SARS-CoV-2, these new strains seem to be also involved in differences in transmission, infectivity, and severity of the disease
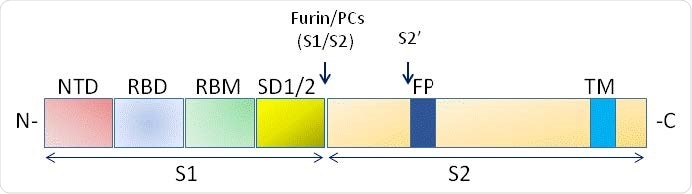
Schematic domain representation of spike glycoprotein, including functional domains in S1 subunit (NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding; SD1/2: subdomain 1 and 2) and in S2 subunit (FP, fusion peptide; TM, transmembrane domain. The N and CT terminal domains are indicated. Arrows denote the protease cleavage sites. PCs: Proprotein convertases.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The viral spike protein
The virus is an enveloped RNA virus with an extensive genome, about 30 kilobases in length. It has four major structural proteins, the Spike (S), Nucleocapsid (N), Envelope (E), and Membrane (M) proteins. The S protein is a transmembrane protein that comprises two subunits, the S1 and the S2, the first mediating receptor binding and the second mediating subsequent fusion between the virus and the cell membrane, as well as cell-cell fusion.
.jpg)
SARS-CoV-2 exploits the angiotensin-converting enzyme 2 (ACE2) to enter target cells. After receptor binding (1), the virus S protein is cleaved by proteases such as furin/TMPRSS2 into S1 and S2 subunits (2) that mediates S2-assisted fusion (3) and the release of the viral genome (4).
The S protein is therefore responsible for the infectivity of the virus and its transmissibility in the host. It binds to the ACE2 molecule via a small folded domain called the receptor-binding domain (RBD). However, this binding is dependent on the cleavage of the spike protein at the S1/S2 junction via a furin-like proprotein convertase in the host cell. Further cleavage of the spike proteins also occurs within the host cells at the S2' site on the S2 domain, which is essential for efficient infection.
The importance of mutations
Viruses, especially RNA viruses, are prone to mutational changes, often with associated changes in viral functions that contribute to increased virulence and mortality. The SARS-CoV-2 has undergone several mutations, with some new variants being VOC-202012/01 (or VUI-202012/01 or B.1.1.7) and 501Y.V2 (or 20C/501Y.V2B.1.351), in the UK and South African variant, respectively.
Both these have mutations at key sites on the RBD, namely, the triplet K417N, E484K, N501Y in the South African variant, and N501Y alone in the British strain.
These variants are rapidly increasing in prevalence, indicating that they have higher transmissibility than the older variants. The current study aimed to understand, via modeling approaches, how ACE2 and spike protein interact and how such interactions contribute to increased pathogenicity and susceptibility to infection.
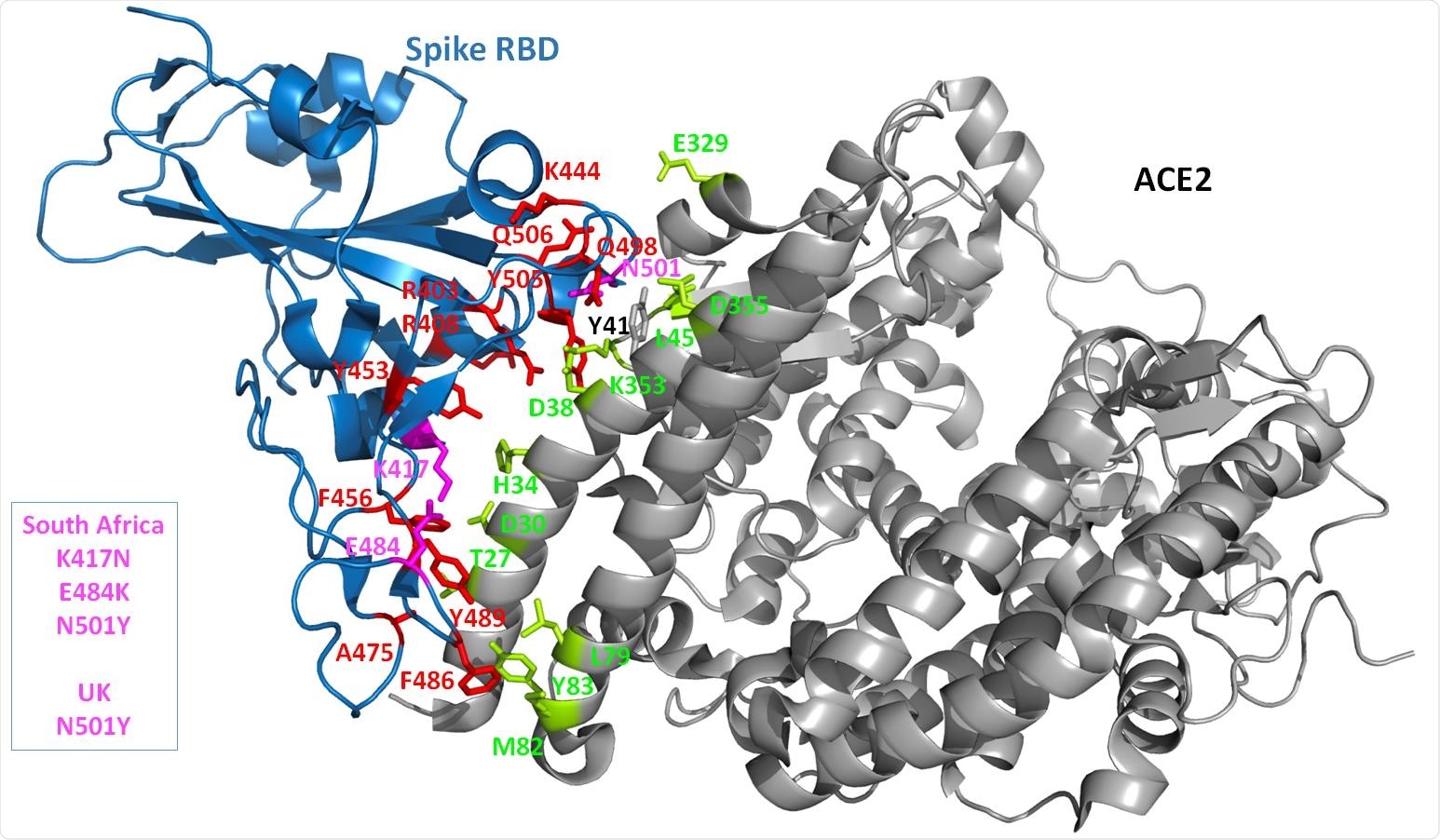
3D structure of the complex. The crystal structure of the Spike RBD and part of the ACE2 receptor interacting with the Spike is shown as cartoon diagram. The Spike domain is in blue while the ACE2 domain is in grey. The Spike side chains of residues predicted to be important (with our in silico protocol, see the Method section) for the interaction with ACE2 are shown in red. The UK and/or South African strains are highlighted by coloring the amino acid changes in this region in magenta. All the Spike side chains shown should have favorable interaction energies with ACE2 but E484 that is predicted to have very weak or unfavorable interactions). On the ACE2 side, the side chains that are predicted to contribute favorably to the interaction with the Spike RBD are shown in lemon. Only ACE Y41 seems to have very limited interactions with the Spike protein while, when the Spike protein carries a Y at position 501, ACE2 Y41 has then some favorable interaction energy values with the Spike.
Study methods
Experimental mutagenesis is a useful tool to investigate the effect of these changes on stability. Such effects can also be estimated using a modeling approach, along with the PPIs between the mutant protein and its receptor or partner. Extant mutagenesis data, confirmed by the current analysis, suggests increased ACE2 affinity with the N501F substitution while leaving its expression intact.
The researchers chose to use multiple computational tools, especially those which can reproduce experimental data, to explore various structural properties of amino acids, including the type of residue, its position, and location with respect to the secondary structure, whether buried or exposed by solvent, and whether at a catalytic site or within a flexible region.
Earlier experimental research has shown that the N501F mutation confers increased binding affinity for the ACE2 on the spike protein. This could not be measured by most modeling platforms used to assess changes in stability at the binding surfaces. For this reason, the current study selected the SPServer and pyDockEneRes tools for their utility in this area.
The researchers compared the results obtained by these methods with experimental results from five protein-protein interactions (PPIs) to assess how well they matched, as well as to understand which substitutions are well-suited to different methods of study.
ACE2 K26 plays minor role
The current study explored specific ACE2 mutations that may contribute to changes in spike binding affinity. The ACE2 S19P mutation, common in Africa, may protect against infection by reducing spike affinity.
An earlier study suggested the K26R mutant, common in Europeans, might increase spike-ACE2 affinity. The researchers in the present study concluded, in contrast, that the K26 residue may not play an important role in modulating spike binding to ACE2 because of its distance from the spike protein in the ACE2-spike complex.
This residue is consistently seen to point away from the spike surface. Even accounting for side-chain flexibility, the distance is too great for hydrogen bonding to occur.
Despite the distance from the spike binding region, a K26 substitution by arginine may allow increased hydrogen bonding, due to the presence of water molecules at the interface. Alternatively, the formation of a salt bridge between the K26 residue and E22, and polar bonds between it and N90, could lead to a local stabilization of this region of the receptor with the K26R mutation, resulting in a more favorable free energy of binding. Further research is required to confirm this finding, which is also supported by other studies.
With the now globally dominant spike D614G variant, an increase in binding affinity is obvious despite this residue's distance from the RBD, being found at the interface between the spike protomers.
Three mutations and their impact
The current study focused on a similar effect resulting from changes in residues present on the RBD and that makes contact with the ACE2 receptor. Crystal structures of both the ACE2 and the RBD have been reported by several earlier studies, aimed at understanding how specific residues act at the interface.
The researchers focused mainly on changes in the amino acids in the two new variants at three sites (K417, E484 and N501), all of which are on the ACE2 binding interface. Such changes can affect protein folding or any of multiple other protein intramolecular and intermolecular interactions.
The N501, found in both strains, and K417, in the South African strain only, affected this interaction, unlike the E484, seen in the latter. The predicted interaction score was less favorable, overall, for the South African strain than for the original structure or for the UK variant.
This N501 residue on the free spike is exposed by solvent but is buried following spike-ACE2 binding. It is in a flexible loop region, and the substitution by a Y does not affect its folding. It could form some hydrogen bonds with K353 of ACE2.
The Y variant in the UK strain has more favorable interaction energy with the receptor. The new side chain allows many favorable interactions with other residues on the ACE2 protein such as pi-stacking, cation-pi interaction, and hydrogen bonding. Such non-covalent interactions could strengthen the bond. Such energetics changes are often difficult to identify in silico.
The K417N mutation also allows proper protein folding but may reduce interaction by abolishing the salt bridge formed with D30 on ACE2, despite some hydrogen bonding potential.
The E484 is on a loop and does not make a significant contribution to binding, probably because it is too far away and cannot form a salt bridge. Moreover, it remains exposed even after ACE2 binding. The replacement by K in the spike protein allows folding but is less favorable for binding, perhaps because it is so near the ACE2 K31 residue.
Thus, all three mutations are less favorable than the N501Y alone, making the predicted ACE2-spike interaction less stable for the South African strain than for the UK strain.
Thus, the study shows that the K417N mutation of the South African strain does not change the expression of the spike but reduces its interaction with ACE2. The E484K mutation also leaves the expression intact, but the K substitution appears to increase binding affinity. Conversely, the modeling platform shows less favorable energetics for K, relative to E, at this site.
This discrepancy could be due to a flawed scoring system or because the interface is more flexible than expected. The latter is difficult to assess in silico. Another explanation could be the effect of long-range interactions between these substitutions and those within the RBD-ACE2 interface.
What are the implications?
Both the UK and South African variants have mutations outside the ACE2-spike binding interface that may affect transmissibility, infectivity, and affinity. At the interface itself, the most important change is the N501Y substitution for the UK strain.
With the South African variant, the presence of the K417N and E484K mutations suggests a reduction in ACE2 binding, the effect of which may or may not compensate for the enhancement due to the N501Y mutation. The observed spread has led scientists to consider this variant as one with potentially greater danger.
If this cannot be explained by increased spike-ACE2 affinity, as the current study indicates, the chief danger of this strain may be due to impaired recognition of the spike protein by neutralizing antibodies as a result of changes at position 484 of the spike, and perhaps other residues. Meanwhile, the increased affinity for ACE2 displayed by the UK strain may confer not just higher infectivity and more rapid transmissibility, but greater virulence.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources