The sudden and devastating emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the etiological agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic – has raised many questions as to where it came from. The possibility of potential animal viral reservoirs, as evidenced by the ready transmission of the virus to mink and back again to mink farmworkers, is also very real. Thus, it is important to understand how this virus arose, and how it is transmitted from its initial animal reservoir to humans.
This virus is one of the SARSr-CoVs among the betacoronaviruses, with 96% whole-genome identity with the bat SC2r-CoV, RaTG13. The latter has therefore been suspected of having given rise to the pandemic virus. Other closely related genomes have been found in pangolins from China and other bats in Eastern China and Japan.
The immediate progenitor has escaped identification, which hampers measures to prevent future outbreaks of the same sort. For this reason, the investigators looked at bats and pangolins to identify SC2r-CoVs, using serological surveys. These, while lacking the definitive nature of polymerase chain reaction (PCR) testing and genome sequencing, are better at identifying infection long after it has resolved.
Identifying new bat- SC2r-CoV in Thailand
The investigators located a colony of 300 bats in an artificial cave in a wildlife sanctuary in eastern Thailand. A third of them were sampled in June 2020. All belonged to the species Rhinolophus acuminatus.
Of the 100 animals, 13 had positive rectal swabs by PCR testing. This showed the presence of a bat CoV that was 96% identical to RaTG13. The most dominant strain of this was named RacCS203, and found to be 94% identical to the bat CoV, RmYN02. RacCS203 is thus a new member of the SC2r-CoV lineage.
It has differences in the ORFs 1ab, 7 and 8, and in part of the nucleocapsid (N) gene, relative to the RmYN02. Orthologous proteins also show different sequences. Moreover, the ORF8 of this strain is closer to that of ZC45 than of RmYN02. The RacCS203 ORF10 is identical to that of the strain ZXC21, but different from that of both the other strains mentioned above.
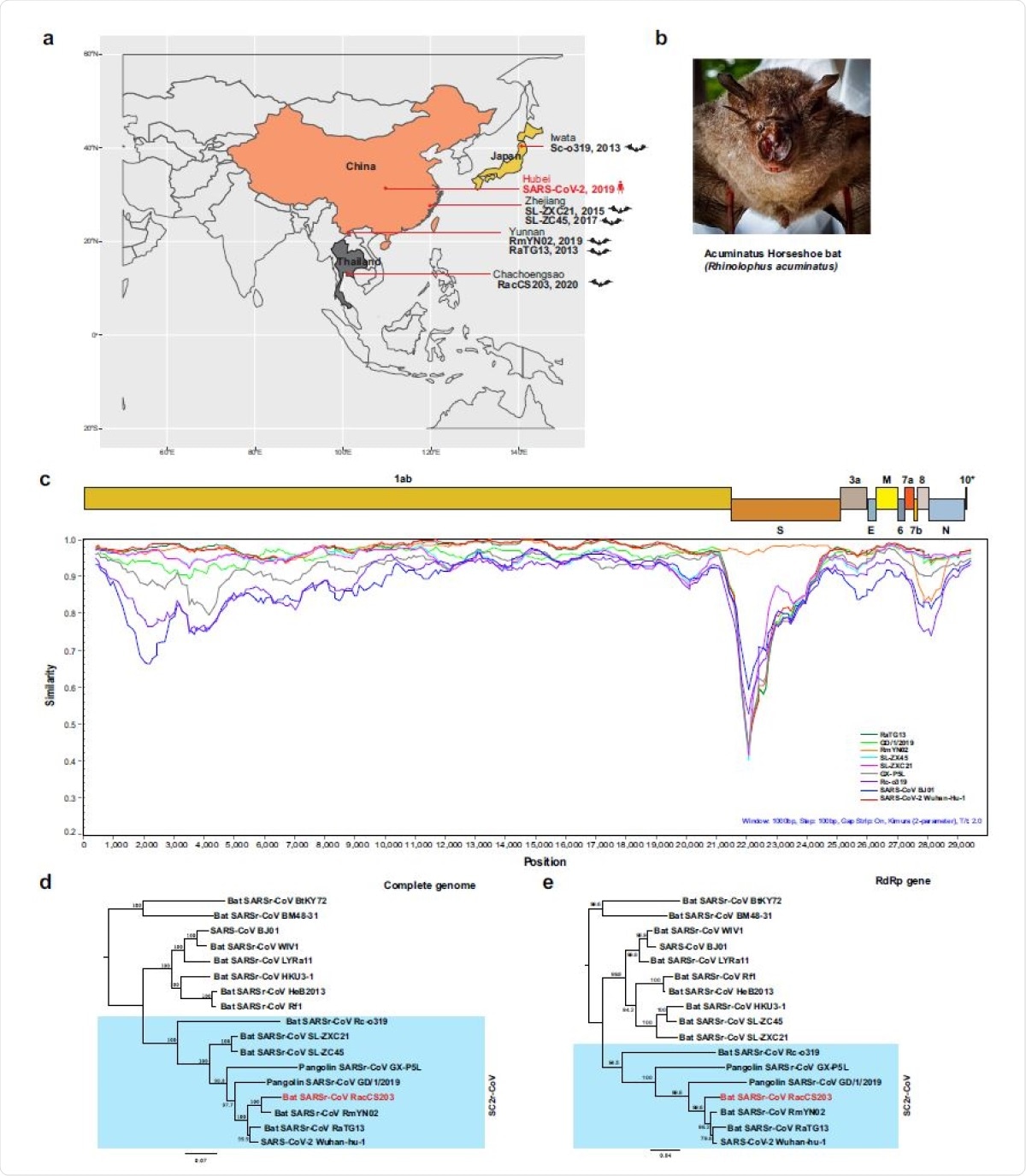
Molecular detection of a SC2r-CoV in bats in Thailand. a Map of Asia illustrating the SC2r-CoVs detected in this region to date. b The Acuminatus horseshoe bats from which the SC2r-CoV was detected. Photo taken by the Thai research team of this study group. c Similarity plot (SimPlot) of wholegenome sequences of 10 SARSr-CoVs using the RacCS203 as a reference genome. d Phylogenetic tree based on whole-genome sequences. e Phylogenetic tree based on the RdRp gene sequences. The trees in d and e were generated using PhyML with general-time-reversible (GTR) substitution model and 1000 bootstrap replicates. Numbers (>70) above or below the branches are percentage bootstrap values for the associated nodes. The scale bar represents the number of substitutions per site. RacCS203 was highlighted in red.
RacCS203 receptor binding domain
ACE2, or the angiotensin-converting enzyme 2 (ACE2), acts as the entry receptor for SARS-CoV-2 in humans. The virus attaches to the receptor via its receptor-binding domain (RBD).
The RacCS203 RBD showed nucleotide sequences that corresponded to non-ACE2-dependent SARSr-CoVs, including RmYN02 and ZC45. This was confirmed experimentally by the lack of ACE2 binding. A pseudovirus expressing the full-length viral spike failed to infect cells in culture when expressing RacCS203 spike, but the successful infection was established with SARS-CoV-2 spike expression.
Serological survey in bats
The researchers used a SARS-CoV-2 surrogate virus neutralization test (sVNT) to explore how far SC2r-CoVs circulated in the region. This test avoided using live SARS-CoV-2 while providing high specificity for antibody-mediated inhibition of RBD-ACE2 binding by this virus.
They found that 4/98 samples had neutralizing antibodies for SARS-CoV-2 RBD-ACE2 binding, with two of them showing 88% and 97% inhibition. This occurred despite the fact that they do not bind ACE2. This inhibition level is surprising, since bat antibodies typically show a lower neutralizing titer than human or rabbit antibodies. In fact, even COVID-19 patients do not show such high titers, for the most part.
The high sequence homology between the RBDs of RacCS203 RBD and SARS-CoV-2 may have elicited cross-neutralizing antibodies without involving ACE2.
Another explanation could be that these reflect cross-neutralization by antibodies induced by other circulating SC2r-CoV that are even more similar to SARS-CoV-2 than RacCS203. In support of this, the PCR positivity was higher among the bats than sVNT, especially for the two samples, which showed high sVNT values.
Pangolin serology
Ten pangolin sera from central and southern Thailand, obtained from confiscated pangolins of unknown country of origin, were also available. The animals all belonged to the species Manis javanica. One of them showed 91% inhibition by the sVNT assay, linking it to a similar result reported from China in another pangolin, 17 years ago.
This shows that these animals can be infected by SC2r-CoVs. They could not be confirmed to be the spillover hosts, however. PCR testing using pan-CoV primers was negative in all samples.
When tested against patient antibody-containing sera from COVID-19 patients, the researchers found that the strongest reactivity was seen with SARS-CoV-2, RaTG13, and GX-P5L, with ZC45 and RmYN02 showing weak reactivity to these antibodies. SARS-CoV showed the least reactivity.
With patient sera from SARS-CoV, low reactivity was observed against all these viruses.
Rabbit sera containing antibodies to RaTG13 RBD were found to be more reactive to the SARS-CoV-2 RBD than when raised against RmYN02 RBD. Both types of sera neutralized SARS-CoV-2, however, by sVNT and by plaque reduction neutralization tests (PRNT).
Geographical range
Rhinolophus bats known to carry SC2r-CoVs are found to occupy a lower latitude than those carrying SARS-CoV-related viruses (SC1r-CoVs). This shows that bats carrying CoVs closely related to SARS-CoV-2 are found to occupy a vast range, extending across multiple regions and countries in Asia, from Japan’s Iwata Prefecture in the north to Chachoengsao, Thailand, in the south; and from Yunnan in the southwest to Zheijang in the east.
So far, five Rhinolophus bat species show the presence of these viruses. Further expansion of these studies, involving different countries, is likely to demonstrate the presence of more SC2r-CoVs in other Rhinolophus species.
What are the implications?
The order of binding affinity to human ACE2 is as follows: GX-P5L>SARS-CoV-2>SARS-CoV>RaTG13. Thus, despite the fact that RaTG13 is the only bat CoV with an RBD that binds hACE2, it has the lowest ranking in order of affinity, while the highest is from the Malayan pangolin. In other words, ACE2 binding does not match with genome sequence identity to SC2r-CoVs or SC1r-CoVs.
These findings suggest that “either these viruses have undergone recombination at the RBD region or ACE2 is not the only or main entry receptor used by these viruses in bats.”
Southeast Asia is likely to harbor many such viruses. The SC2r-CoV lineage is also capable of great diversity, showing multiple mutations unique to each of the viral lineages in this group. This indicates “that there is a high probability to find the immediate progenitor virus of SARS-CoV-2 with intensified and internationally coordinated surveillance.”