Since the start of the COVID-19 (coronavirus disease 2019) pandemic in late December 2019, the etiological agent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) of this disease has caused more than 2.6 million deaths worldwide from over 117 million reported infections. However, as new vaccines are administered against the ancestral strain, many mutated variants of the virus are emerging across the world.
To develop complementary strategies urgently needed to avert any viral escape, researchers are using alternative approaches. Nanobodies are drawing attention recently as potent therapeutic alternatives.
In a recent bioRxiv* preprint paper, researchers isolated anti-RBD nanobodies from llamas and “nanomice” (they engineered to produce VHHs cloned from alpacas, dromedaries and camels). In the study, they identified two sets of highly neutralizing nanobodies - one that recognized a region outside the ACE2-binding site, and the other exclusively focused on the RBD (receptor binding domain)-ACE2 (angiotensin-converting enzyme 2) interface, failing to neutralize pseudoviruses carrying the E484 or N501Y substitutions.
The SARS-CoV-2 envelope is coronated with the glycoprotein Spike, enabling the virus’s entry into the host human cells. The S protein binds to the ACE2 on the host cells via the RBD.
The high affinity of RBD to bind to the ACE2 underlies the higher transmissibility of SARS-CoV-2. Therefore, this region is the target of designing effective antibodies and vaccines.
Nanobodies are tiny, antigen-binding proteins. These are variable domains (VHH fragments) of heavy chain-only antibodies (HCAbs) isolated from camelids. Compared to mouse and human antibodies (~150 kDa in size), the nanobodies are the variable domains of camelids heavy chain-only antibodies retaining full antigen specificity at a molecular weight of ~15 kDa.
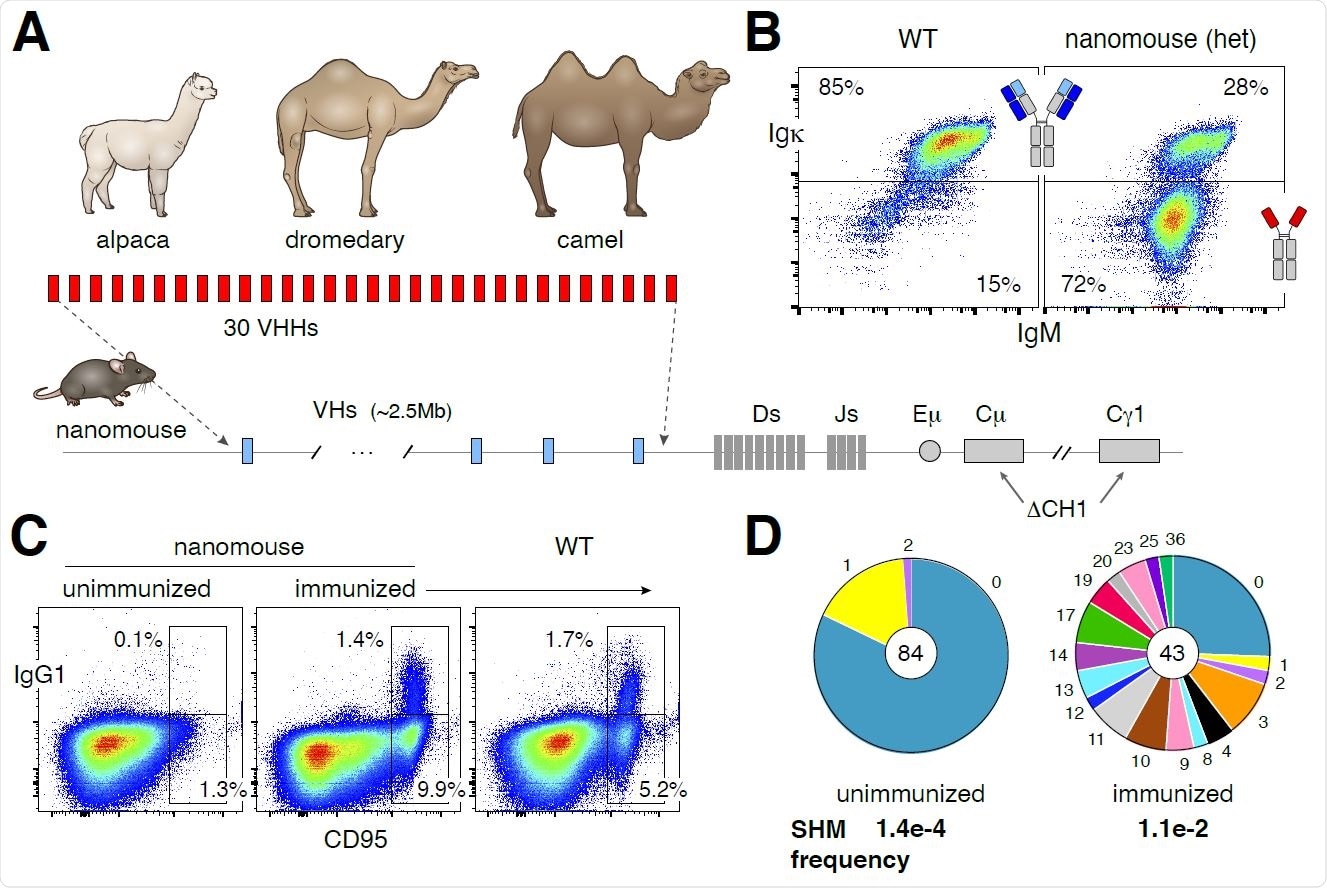
Production and characterization of nanomice. (A) 30 VHHs were selected from alpaca, dromedary and camel to create a cassette, which was inserted via CRISPR-Cas9 in lieu of the mouse VH locus (2.5MB in size). The CH1 exon of Cμ and Cg1 was also deleted to avoid misfolding of the antibody heavy chain when unable to pair with light chains. (B) Flow cytometry analysis of splenic B220+ B cells from WT or heterozygous nanomice. IgM+Igk+ represent cells expressing conventional heavy-light chain antibodies, whereas IgM+Igk- are mostly Igl+ in WT (not shown) or single-chain antibody B cells in nanomice. (C) Flow cytometry analysis of splenic cells from unimmunized and immunized nanomice and controls stained with the germinal center markers CD95 and IgG1. (D) Pie charts showing VHH somatic hypermutation in unimmunized and immunized nanomice. Pie segments are proportional to the VHH sequences carrying the mutations indicated on the periphery of the chart. The middle circle contains the total number of sequences and the mutation frequency is shown in bold below.
Because of their small size, the nanobodies can recognize protein crevices that are inaccessible to conventional antibodies. The nanobodies have high affinity, easier to use and produce. Nanobodies represent promising tools to prevent COVID-19 mortality when vaccines are compromised, the researchers stated.
Despite the advantages of the nanobodies, these are not popularly used because 1) camelids are large animals and therefore expensive to maintain, especially in academic facilities, and 2) highly heterogeneous immune responses are found (due to lack of inbreeding). High specificity and diversity are difficult to achieve by other means.
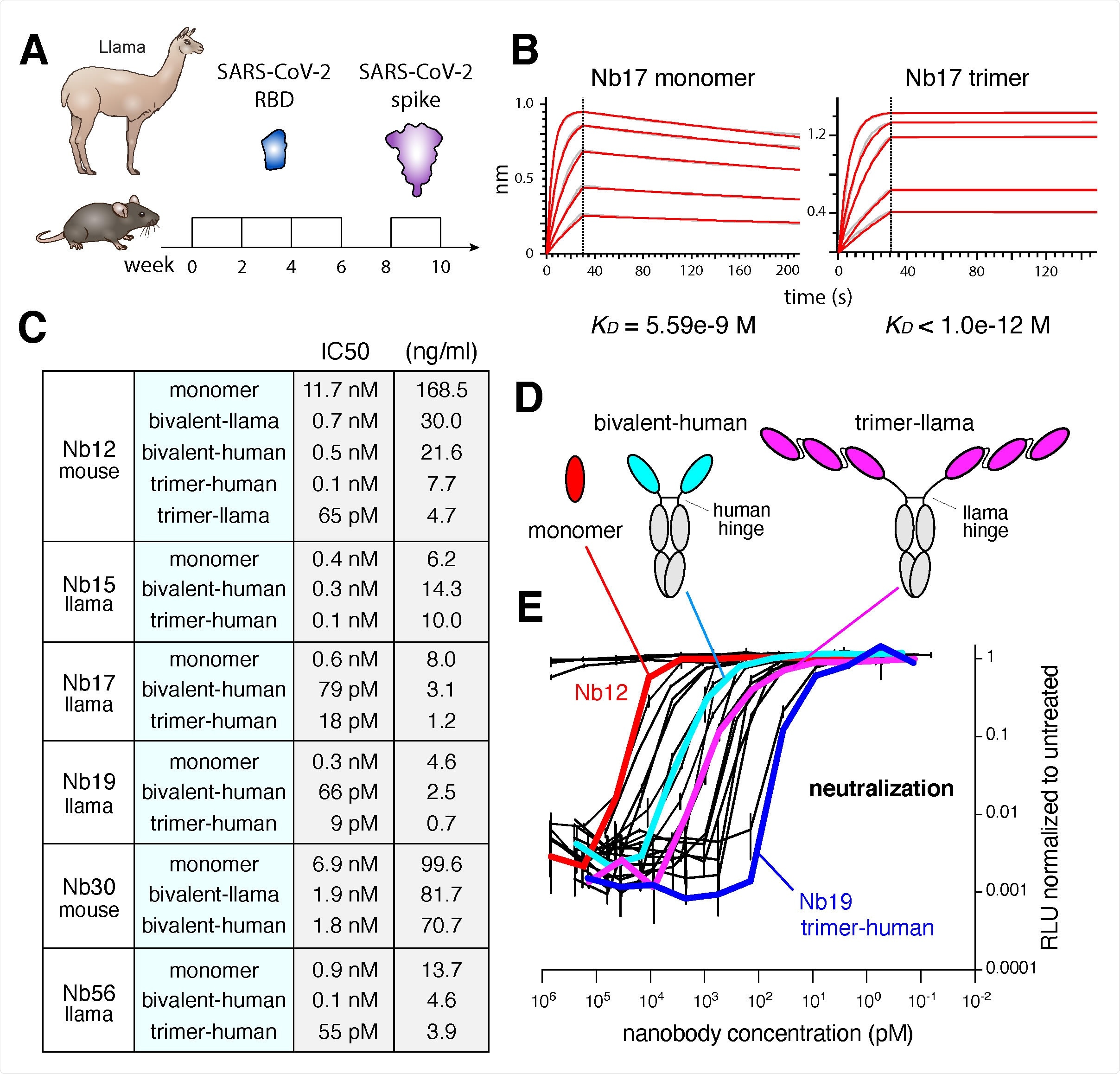
Isolation of Nbs against SARS-CoV-2. (A) Immunization regimen to obtain high affinity Nbs against the SARS-CoV-2 RBD from llama and nanomice. (B) BLI analysis of difference concentrations of Nb17 monomer and trimer binding to immobilized RBD. Red trace represents the raw data, and the kinetic fit is shown in grey underneath. Equilibrium (KD) constants are provided. (C) Table summarizing pseudovirus neutralization potency (IC50) of selected Nbs. Values are provided in molarity or as ng/ml. (D) Diagrams showing Nbs used in neutralization assays as monomers, bivalent or trimers (the last two fused to human IgG Fc via the human or llama hinge domain. (E) Neutralization of SARS-CoV-2 pseudovirus (luciferase assay) by the 20 Nb variants shown in panel C. Nb12 as monomer (red), pair (cyan) and trimer (magenta), as well as Nb19 as trimer (blue) are highlighted.
Therefore, in this study, the researchers produced the Nbs in mice by combining 18 alpaca, 7 dromedary, and 5 camel VHH genes in a 25Kb insertion cassette. They fused each gene to a functional mouse VH promoter, leader exons, and a downstream recombination signal sequence (RSS) to ensure physiological expression and recombination.
“By means of CRISPR-Cas9 and recombinase-mediated cassette exchange (RMCE), the VHH cassette was inserted in lieu of the entire VH locus in mouse ES cells.”
Here, the mouse chimeras were derived from ES cells, and the targeted allele was germline transmitted to F1 offspring, which they referred to as nanomice in the study.
The research found that a large fraction of nanomouse B cells developed expressing single-chain antibodies. They concluded that all VHH genes in nanomice undergo V(D)J recombination and are thus potentially available for expansion during the adaptive immune response.
Based on their results, they also concluded that mouse B cells could mature, expressing single-chain antibodies. They found excellent B cell activation and affinity maturation in the nanomice.
The researchers also tested the multimeric constructs of these nanobodies and found remarkable avidity - the best reported to date for anti-SARS-CoV2 Nbs. They also reported that these multimeric nanobodies efficiently overcame the SARS-CoV-2 escape mutants.
To obtain a structural understanding of the neutralizing regions recognized by nanomouse and llama Nbs, the researchers conducted a 3D electron microscopic study of the nanobodies in complex with the Spike protein. These reconstructions revealed the llama nanobodies uniformly target the ACE2-binding interface, whereas the nanomouse nanobodies recognize the RBD at a surface outside the ACE2-binding site.
In conclusion, the researchers highlighted their study’s essential contribution to the creation of Nb-producing mice. Notably, the researchers emphasize that if this model ultimately replaces the use of camelids in biomedical research, then the llamas and alpacas are protected, which otherwise are invariably culled in antibody production farms after a given study.
They stated in their study that as proof of principle, we used the nanomice to produce highly specific nanobodies against the SARS-CoV-2 RBD. Due to their versatility and capacity for multivalency, these anti-RBD nanobodies can overcome possible mutations occurring at or around the ACE2 binding motif. This study has isolated and characterized two sets of nanobodies that effectively neutralize pseudotyped viruses carrying RBD mutations from U.K. and South African-New York variants.
“The Nbs described here solve the challenge posed by SARS-CoV-2 escape variants either through enhanced avidity as homotrimeric constructs or by binding conserved epitopes typically unavailable to human monoclonal antibodies.”
Journal reference:
- Multimeric nanobodies from camelid engineered mice and llamas potently neutralize SARS-CoV-2 variants, Jianliang Xu, Kai Xu, Seolkyoung Jung, Andrea Conte, Jenna Lieberman, Frauke Muecksch, Julio Cesar Cetrulo Lorenzi, Solji Park, Zijun Wang, Lino Tessarollo, Tatsiana Bylund, Gwo-Yu Chuang, Adam S. Olia, Tyler Stephens, I-Ting Teng, Yaroslav Tsybovsky, Tongqing Zhou, Theodora Hatziioannou, Paul D. Bieniasz, Michel C. Nussenzweig, Peter D. Kwong, Rafael Casellas, bioRxiv, 2021.03.04.433768; doi: https://doi.org/10.1101/2021.03.04.433768, https://www.biorxiv.org/content/10.1101/2021.03.04.433768v1