Although several vaccines have now been approved to combat COVID-19, not enough is known about their efficacy against new variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Available treatments target the life-cycle of the virus or immune response of the host. Other possible targets are the angiotensin-converting enzyme 2 (ACE2) or the protease TMPRSS2, which are key during the initial virus infection stages. Antiviral drugs are another class of drugs being investigated.
Developing new drugs is expensive and takes a long time. So people have been looking to repurpose drugs that have already been approved. Rather than experimentally screening drugs, computational methods can be faster and would take lesser time and resources.
In a study published in the bioRxiv* preprint server, researchers propose an approach for prioritizing drug candidates for treating COVID-19. The team selected compounds from the DrugBank library by virtual screening using cheminformatics and bioinformatics approaches for potential antivirals against COVID-19 and experimentally tested the performance of the top candidates in vitro.
Screening for potential drugs
The authors analyzed publicly available transcriptomics data to determine the action mechanism of drugs and changes made according to specific diseases. They identified structures in the compounds that can affect initial viral infection, revert any virus-induced transcriptome changes, and target genes in host immune response. In addition, they also selected chemical structures that were previously identified as active against SARS-CoV-2.
Based on these criteria, the team selected 700 candidates for further study. They also included criteria such as drug availability, price, and storage in their analysis. From the initial list, they further shortened it to 23 to experimentally test.
About half of these relate to treatments of tumors, and eight are kinase inhibitors. Kinases are important in many biological processes and have been proposed as COVID-19 treatments. The drugs selected comprise antivirals, antimicrobials, and anticancer treatments.
About half of the drugs identified using the approach are already being investigated for COVID-19 treatment. This demonstrates the capability of the method to identify potential drug candidates and also identify others that have not been used in association with COVID-19.
The team tested the effect of the selected drug candidates on HEK-293T cells expressing human ACE2 and TMPRSS2. Two drugs, 7-hydroxystaurosporine and bafetinib, showed significant inhibition of virus-infected cells.
7-Hydroxystaurosporine is an antitumor drug used for treating cancers such as leukemia. But, it has not been associated with COVID-19 yet. Bafetinib is a tyrosine kinase inhibitor used against leukemia and was also identified as a SARS-CoV-2 inhibitor in other studies.
The authors tested if a combination of the two would be better in inhibiting SARS-CoV-2. They found that 7-hydroxystaurosporine alone had significant inhibition of viral infection at low concentrations that are not cytotoxic. A combination with bafetinib reduced the number of infected cells by more than 80%, with no cytotoxic effects. Further testing revealed that the drugs act by inhibiting the virus after it has entered the cells.
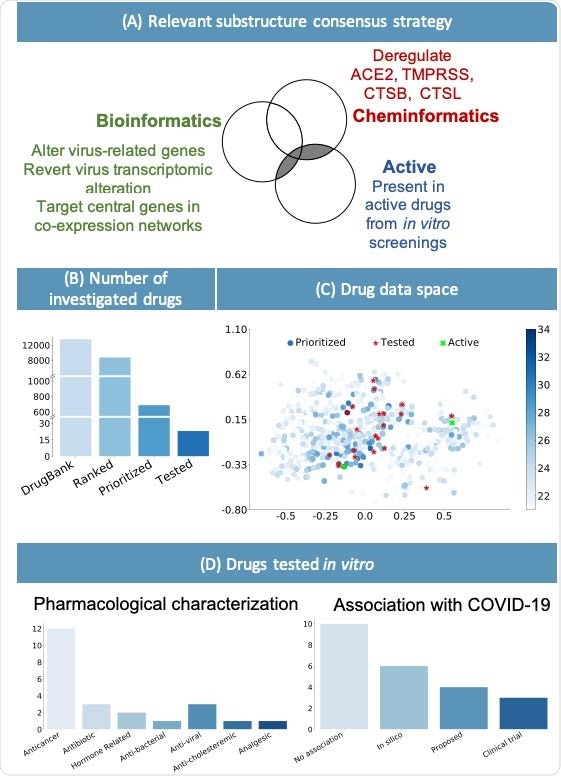
(A) Consensus strategy to identify relevant chemical substructure, using bioinformatics and cheminformatics methods as well as experimental results from published literature. (B) The suggested approach allows reducing the number of experimental tests: the whole DrugBank database was filtered to less than 2000 relevant drugs and in vitro testing was performed on 23 candidates. (C) Graphical representation of the prioritized drugs. The shade of blue represents the number of chemical substructures identified in (A), present in the drugs. The 23 selected compounds are shown in red. They were selected among the drugs sharing the most relevant substructure as well as satisfying practical logistic criteria. Of the 23 drugs, the 2 highlighted in green have been experimentally identified as active. (D) Pharmacological characterization and description of known association with COVID-19 of the 23 tested drugs. In silico refers to drugs derived from in silico studies, while proposed refers to drugs suggested for their potential therapeutic role in literature.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Possible mechanism of action
Some human pathogenic viruses like SARS-CoV-2 can cause cell-to-cell fusion, known as syncytia formation, which has been associated with the viral spread and tissue damage. SARS-CoV-2 infection of HEK-293T cells leads to their fusion, forming large multinucleated cells.
Image analysis showed 7-hydroxystaurosporine could inhibit this cell-to-cell fusion, and the combination with bafetinib increased this effect. Bafetinib is known to increase the accumulation of anticancer drugs in cells, and this could provide a mechanism of its inhibitory action when combined with 7-hydroxystaurosporine.
The researchers also analyzed the chemical structures of the two drugs. Both are highly aromatic with a conjugated π-bond system and have a planar geometry. Their 3-D geometries were about 67% similar, which could explain why they both have antiviral activity.
Models of bafetinib have shown it can block an efflux transporter and that could explain its inhibitory effect. But, since both have inhibitory effects alone, the exact mechanism of how this happens is still not clear.
“The prioritization methodology we developed allowed us to make an informed decision about which drugs to test, out of almost 8,000 drugs from DrugBank, thus significantly reducing the costs of drug repurposing, while simultaneously increasing the success rate,” write the authors. This approach could help to repurpose of drugs not only for viral infections but also for others.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Serra, A. et al. (2021) Computationally prioritized drugs inhibit SARS-CoV-2 infection and syncytia formation. bioRxiv, https://doi.org/10.1101/2021.04.15.440004, https://www.biorxiv.org/content/10.1101/2021.04.15.440004v1
- Peer reviewed and published scientific report.
Serra, Angela, Michele Fratello, Antonio Federico, Ravi Ojha, Riccardo Provenzani, Ervin Tasnadi, Luca Cattelani, Giusy del Giudice, Pia A S Kinaret, Laura A Saarimäki, Alisa Pavel, Suvi Kuivanen, Vincenzo Cerullo, Olli Vapalahti, Peter Horvath, Antonio Di Lieto, Jari Yli-Kauhaluoma, Giuseppe Balistreri, Dario Greco. 2021. “OUP Accepted Manuscript.” Briefings in Bioinformatics. https://doi.org/10.1093/bib/bbab507. https://academic.oup.com/bib/article/23/1/bbab507/6484515.