Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative pathogen of the coronavirus 2019 (COVID-19) pandemic. To date, over 170 million cases have been confirmed worldwide, resulting in over 3.55 million deaths.
While several vaccine candidates have been developed and deployed to arrest the spread of SARS-CoV-2, concern among scientists and public health authorities is mounting over the immunity escape potential of emerging viral variants.
To address this problem, a group of scientists in Germany has undertaken research to identify molecular targets across viral variants to aid in the development of second and third-generation vaccines and novel therapeutics.
The study identifies residues across the variants which are significant for viral binding to the host cell, which can then be used as primary targets for intervention by neutralizing antibodies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
How does SARS-CoV-2 infect host cells?
To infect a cell, SARS-CoV-2 interacts with the cell surface protein angiotensin-converting enzyme 2 (ACE2) through its spike protein. Within the spike (S) protein, the receptor-binding domain (RBD) interfaces with ACE2 enabling the virus to bind to the host cell surface.
The host cell transmembrane, serine protease TMPRSS2, then allows the spike protein to be primed, releasing the fusion peptide from the protein backbone, which, after insertion into the host cell membrane and the activation of a conformational switch, leads to the dissociation of the spike protein’s S1 and S2 subunit domains. This then allows the virus and host cell membrane to fuse, and all non-cleaved subunits of the spike protein dissociate from the ACE2 receptors.
This simplified view of the life cycle of SARS-CoV-2 illustrates the importance of optimized binding efficiency between the interaction of RBD and ACE2 for the virus to enter the host cell. Research into variants has shown that there are amino acid exchanges at the RBD and ACE2 interface in more infectious variants compared to the wild-type.
The study
The researchers analyzed RBD variants in complex with ACE2 in the following confirmed variations of the virus: B.1.1.7 (from the UK), B.1.1.7+E484K (from the UK), B.1.351 (from South Africa) and P.1 (from Brazil). They were able to use molecular dynamics (MD) simulations in order to observe and examine these variants effectively.
The study showed that progressive viral variants can occur with reduced binding energies to ACE2. The scientists uncovered how the decomposition of the RBD-ACE2 interface could help identify residues on the RBD which are significant for the virus to interact with the host cell in different viral variants of this infection. They found critical amino acids, including phenylalanine 486, glutamine 493 and tyrosine 505, as important residues in the RBD-ACE2 complex in all the RBD variants analyzed.
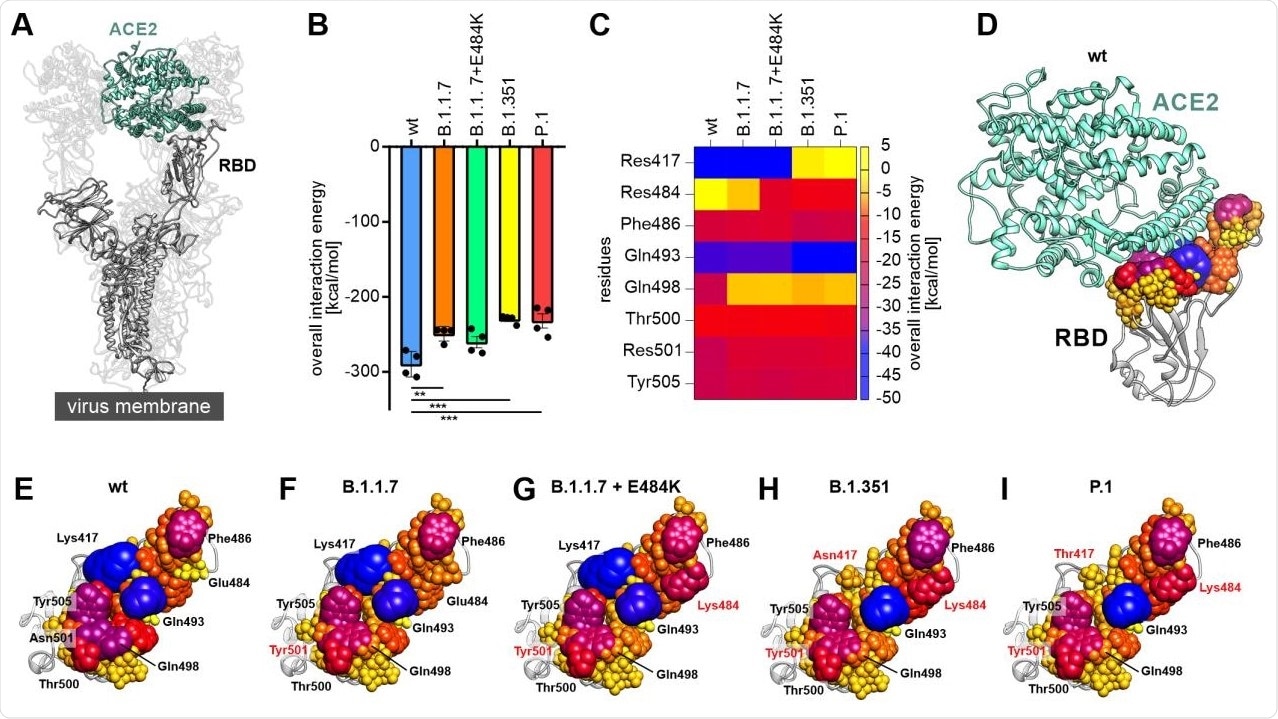
Decomposition of the overall linear interaction energies. (A) Structural representation of the trimeric spike protein as expressed on the viral membrane (grey) with exposed receptor binding domain (RBD). The host cell receptor ACE2 is displayed in aquamarine (PDB ID: 7kms (19)). (B) Overall linear interaction energy as calculated for the different spike protein variants (wt = wild type). Statistical analysis was performed using one-way ANOVA (n=4, differences assumed significant for ** p<0.01, *** p<0.001). (C) Heat map of receptor-binding domain (RBD) expressed residues important for binding of ACE2. (D) Wild type RBD in complex with ACE2 (aquamarine). All residues within a maximum distance of 8 Å to ACE2 are displayed according to their interaction energy with different colors and sphere radii. (E) View on the interface formed by the wild type RBD with residues displayed in different colors and sphere diameters according to their interaction energy. Especially lysine 417 (Lys417), glutamate 484 (Glu484), phenylalanine 486 (Phe486), glutamine 493 (Gln493), glutamine 498 (Gln498), threonine 500 (Thr500), asparagine 501 (Asn501) and tyrosine 505 (Tyr505) were of interest. (F) View on the interface formed by the RBD of the B.1.1.7 variant with ACE2. Note that residue 501 is mutated to tyrosine (Tyr501; red) in this variant. (G) View on the interface formed by the RBD of the B.1.1.7+E484K variant with ACE2. Note the additional change of residue 484 from glutamate to lysine (Lys484; red) compared to wild type and B.1.1.7. (H) View on the interface formed by the RBD of the B.1.351 variant with ACE2. This variant carries the tyrosine at position 501, the lysine at position 484 and an additional exchange from lysine to asparagine at position 417 (Asn417; red). (I) View on the interface formed by the RBD of the P.1 variant with ACE2. Besides a tyrosine at position 501 and a lysine at position 484 this variant carries a lysine to threonine exchange at position 417 (Thr417; red).
Significance for COVID-19 pandemic and research
The researchers analyzed RBD variants in four different confirmed variants across the globe and investigated the relationship between RBD and ACE2. In doing so, they were able to identify epitopes that are commonly found in all variants. These epitopes can possibly be used as molecular targets for neutralizing antibodies and other therapeutics to be developed to prevent viral entry and limit the severity of SARS-CoV-2 infections overall.
The use of molecular dynamics simulations allowed the researchers to track the changes between the interaction energies of RBD and ACE2, which ultimately led them to uncover that there was a trend in the variants towards having reduced interaction energies between the two. This indicated that the ‘binding’, as well as the dissociation of the virus from the ACE2 receptor, could be a driving force for viral evolution, resulting in the emergence of novel variants.
The team’s research provides good ground for the future development of therapeutics that are cross-reactive against novel SARS-CoV-2 variants that have exhibited immunity escape potential via both vaccination and natural infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Socher, E., Conrad, M., Heger, L., Paulsen, F., Sticht, H., Zunke, F. and Arnold, P., 2021. Decomposition of the SARS-CoV-2-ACE2 interface reveals a common trend among emerging viral variants. doi: https://doi.org/10.1101/2021.05.28.446149, https://www.biorxiv.org/content/10.1101/2021.05.28.446149v1
- Peer reviewed and published scientific report.
Socher, Eileen, Marcus Conrad, Lukas Heger, Friedrich Paulsen, Heinrich Sticht, Friederike Zunke, and Philipp Arnold. 2021. “Computational Decomposition Reveals Reshaping of the SARS‐CoV‐2–ACE2 Interface among Viral Variants Expressing the N501Y Mutation.” Journal of Cellular Biochemistry 122 (12): 1863–72. https://doi.org/10.1002/jcb.30142. https://onlinelibrary.wiley.com/doi/10.1002/jcb.30142.