Researchers in the United States have provided a high-resolution description of the viral dynamics involved in acute infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The team’s daily longitudinal sampling of 60 newly infected individuals revealed that significant person-to-person variation in the shedding of infectious virus contributes to superspreading.
Superspreading is the term used to describe a small fraction of infected individuals accounting for a disproportionate level of community transmission.
“These results provide the first high-resolution, multi-parameter empirical profile of acute SARS-CoV-2 infection in humans and implicate person-to-person variation in infectious virus shedding in driving patterns of epidemiological spread of the pandemic,” writes Christopher Brooke from the University of Illinois at Urbana-Champaign and colleagues.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes pee review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The dynamics of SARS-CoV-2 shedding is poorly understood
“Numerous behavioral and environmental explanations have been offered to explain transmission heterogeneity, but the extent to which the underlying features of the infection process within individual hosts contribute towards the superspreading phenomenon remains unclear,” says Brooke and the team.
The researchers say that addressing this knowledge gap will inform the design of more targeted and effective strategies for controlling community spread.
What did the researchers do?
The team performed a daily longitudinal sampling of sixty SARS-CoV-2-infected individuals for 14 days to capture the dynamics of infectious virus and viral RNA shedding during acute infection.
By fitting mechanistic models, the team directly estimated viral reproduction, clearance and infectiousness for each individual.
During the Fall of 2020 and the Spring of 2021, all faculty, staff, and students at the University of Illinois at Urbana-Champaign underwent reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for SARS-CoV-2 at least twice a week.
Brooke and colleagues leveraged this large-scale screening to enroll infected individuals (median age 28 years; age range 19 to 73 years) and tested daily nasal and saliva samples to generate a high-resolution portrait of viral dynamics during the early stage of infection.
What did they find?
The results revealed a high level of person-to-person variation in the shedding of infectious virus, which the team says provides a partial explanation for the role that super spreaders play in the community transmission of SARS-CoV-2.
The study revealed significant individual-level differences in the duration of infectious virus shedding, in clearance kinetics, and in the temporal relationship between shedding in nasal and saliva compartments.
When the researchers compared the reported symptom frequencies for days where individuals tested culture-positive or negative for infection, muscle aches, runny nose, and scratchy throat were significantly more likely to be reported on days when participants tested positive.
This suggests that these specific symptoms could serve as potential indicators of infectious status, say the researchers.
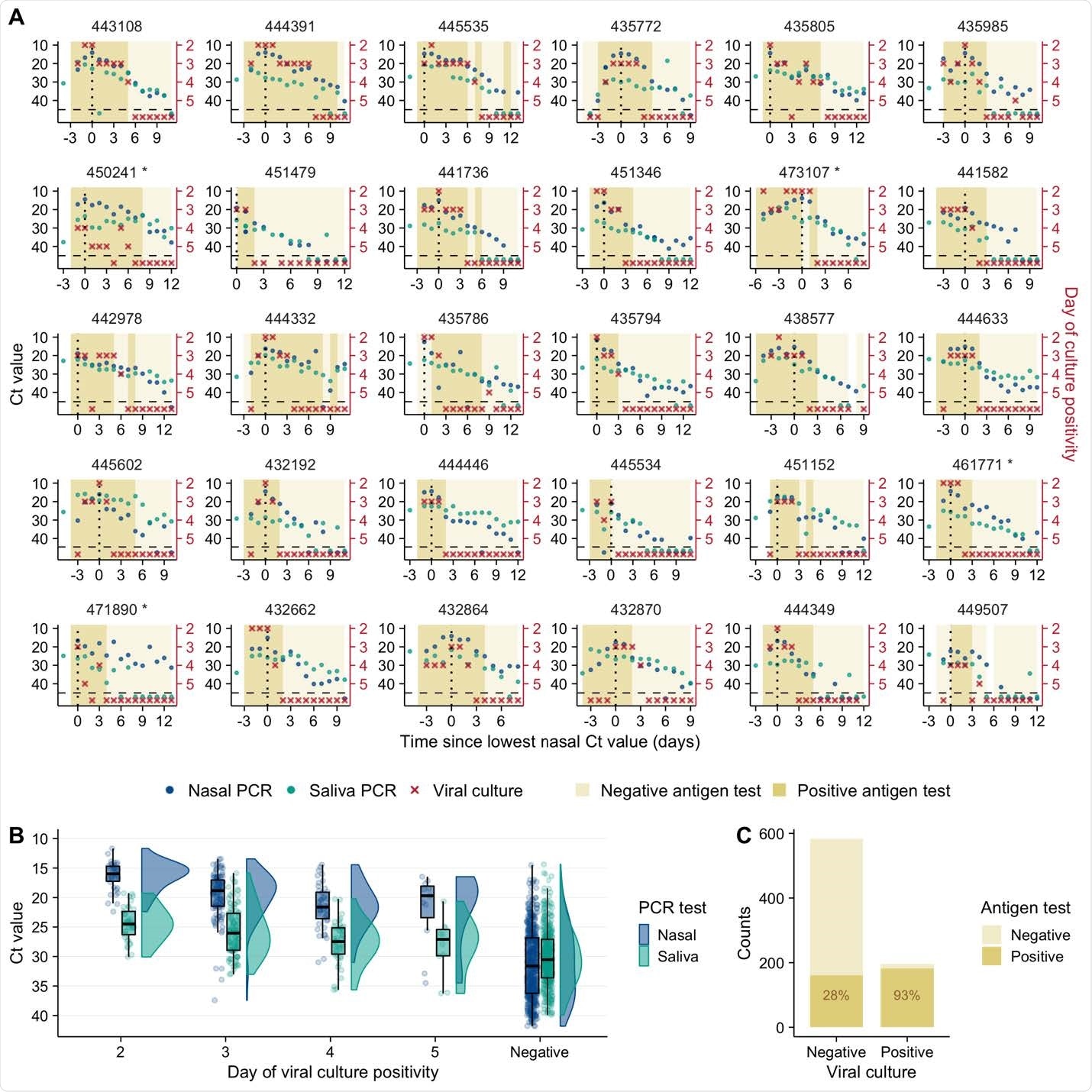
SARS-CoV-2 viral dynamics captured through daily sampling. (A) Temporal trends for the saliva RTqPCR (teal dots), nasal swab RTqPCR (navy blue dots), nasal swab viral culture (red crosses), and positive nasal swab antigen test results (dark mustard shaded area). The X axis shows days since the measured peak in nasal swab viral genome load, which is indicated by a vertical dotted line. The left Y axis indicates Ct values for saliva RTqPCR assay (covidSHIELD) and Cn values for nasal swab RTqPCR assay (Abbott Alinity). The right Y axis indicates results of viral culture assays, where day of culture positivity indicates the day of incubation at which > 50% of Vero-TMPRSS2 cells infected with the sample were positive for cytopathic effect. Horizontal dashed line indicates limit of detection of RTqPCR and viral culture assays. Title of each plot indicates the participant ID for the top 30 individuals with the most data points (the remaining 30 participants are shown in figure S1). Single asterisk next to the participant ID indicates B.1.1.7 variant infection. (B) Individual Ct (for saliva) and Cn (for nasal swabs) values from samples plotted based on concurrent results from the viral culture assay.
Viral genome shedding peaked earlier in saliva samples
Brooke and colleagues observed a surprising degree of discordance in viral dynamics between nasal and saliva samples among most participants.
Among 41 of 54 individuals analyzed, viral genome shedding peaked at least one day earlier in saliva samples than in nasal samples.
The team says this indicates strong compartmentalization of the nasal and oral cavities and suggests that saliva could serve as a superior sampling site for the early detection of infection.
In addition, viral clearance differed significantly between nasal and saliva samples, with the post-peak viral genome load declining more rapidly in nasal samples than in saliva samples.
Individual-level infectiousness was also highly variable, with a more than 30-fold difference between the highest and lowest estimated infectiousness.
“This emphasizes the potential for a small subset of individuals that exhibit high intrinsic infectiousness to function as ‘superspreaders’ if they have frequent and/or high-risk contacts during the infectious period,” write the researchers.
Comparison of B.1.17 (alpha) lineage and non-B.1.17 viruses
The researchers compared shedding dynamics between the B.1.1.7 (alpha) SARS-CoV-2 variant and non-B.1.1.7 viruses to investigate whether any significant shedding differences could explain the enhanced transmissibility of B.1.1.7.
Analysis of 15 individuals infected with B.1.1.7 suggested that viral genome shedding dynamics in nasal samples are indistinguishable between B.1.1.7 and non-B.1.1.7 infections and that there was no significant difference in overall infectiousness between B.1.1.7 and non-B.1.1.7 viruses.
However, analysis of saliva samples revealed a slower rise in pre-peak viral loads for B.1.1.7, compared with non-B.1.1.7 viruses.
“Because symptom onset seems to occur around the time of peak viral load, the prolonged pre-peak shedding in saliva by B.1.1.7 could potentially enhance transmissibility by extending the pre-symptomatic shedding period of the virus,” suggests the team. “However, more data are needed to determine whether there is a mechanistic link between pre-peak shedding kinetics and transmission potential.”
What did the authors conclude?
Brooke and colleagues say the results of this daily multi-compartment sampling of viral dynamics among dozens of individuals newly infected with SARS-CoV-2 provides the most comprehensive, high-resolution description of SARS-CoV-2 shedding and clearance dynamics in humans to date.
“Altogether, our data provide an unprecedented view of the longitudinal viral dynamics of SARS-CoV-2 infection in humans and implicate both individual-level heterogeneity in viral shedding and the oral compartment and as playing critical roles in transmission,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Brooke C, et al. Daily sampling of early SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. medRxiv, 2021. doi: https://doi.org/10.1101/2021.07.12.21260208, https://www.medrxiv.org/content/10.1101/2021.07.12.21260208v1
- Peer reviewed and published scientific report.
Ke, Ruian, Pamela P. Martinez, Rebecca L. Smith, Laura L. Gibson, Agha Mirza, Madison Conte, Nicholas Gallagher, et al. 2022. “Daily Longitudinal Sampling of SARS-CoV-2 Infection Reveals Substantial Heterogeneity in Infectiousness.” Nature Microbiology 7 (5): 640–52. https://doi.org/10.1038/s41564-022-01105-z. https://www.nature.com/articles/s41564-022-01105-z.