Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a novel coronavirus that has caused infections and deaths in millions of people worldwide since its emergence in Wuhan, China, in late December 2019.
SARS-CoV-2 affects not only humans but also cats, dogs, ferrets, hamsters, and non-human primates. The virus was even found to affect minks, and cases of mink-to-human cross-species transmission have been reported.
"The spike (S) glycoprotein of SARS-CoV-2 is the main determinant of host tropism and susceptibility, and the main target of antibody responses", says the team.
Therefore, the emergence of adaptive mutations in the spike protein has a strong effect on host tropism and viral transmission.
The spike protein comprises of two subunits: S1 subunit having the receptor-binding domain (RBD) that helps it to bind to the angiotensin-converting enzyme 2 (ACE2), which is present on the cell surface of the host, and the S2 subunit that helps in the fusion of the cellular membrane and the viral membrane.
"To fuse with the host cell, the S protein needs to be cleaved by cellular proteases at the S1/S2 and S2' sites", says the team.
The S1/S2 site consists of a multibasic furin motif that can undergo processing by the furin proteases or by transmembrane serine proteases.
Since 2019, several SARS-CoV-2 lineages termed as Variants of Concern (VOC) have emerged that have increased the virus's transmissibility. These variants have arisen mainly due to spike protein mutations.
In this study, the researchers characterized the spike polymorphisms of SARS-CoV-2 both in vitro and in vivo in order to understand viral pathogenicity, transmissibility, and fitness.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
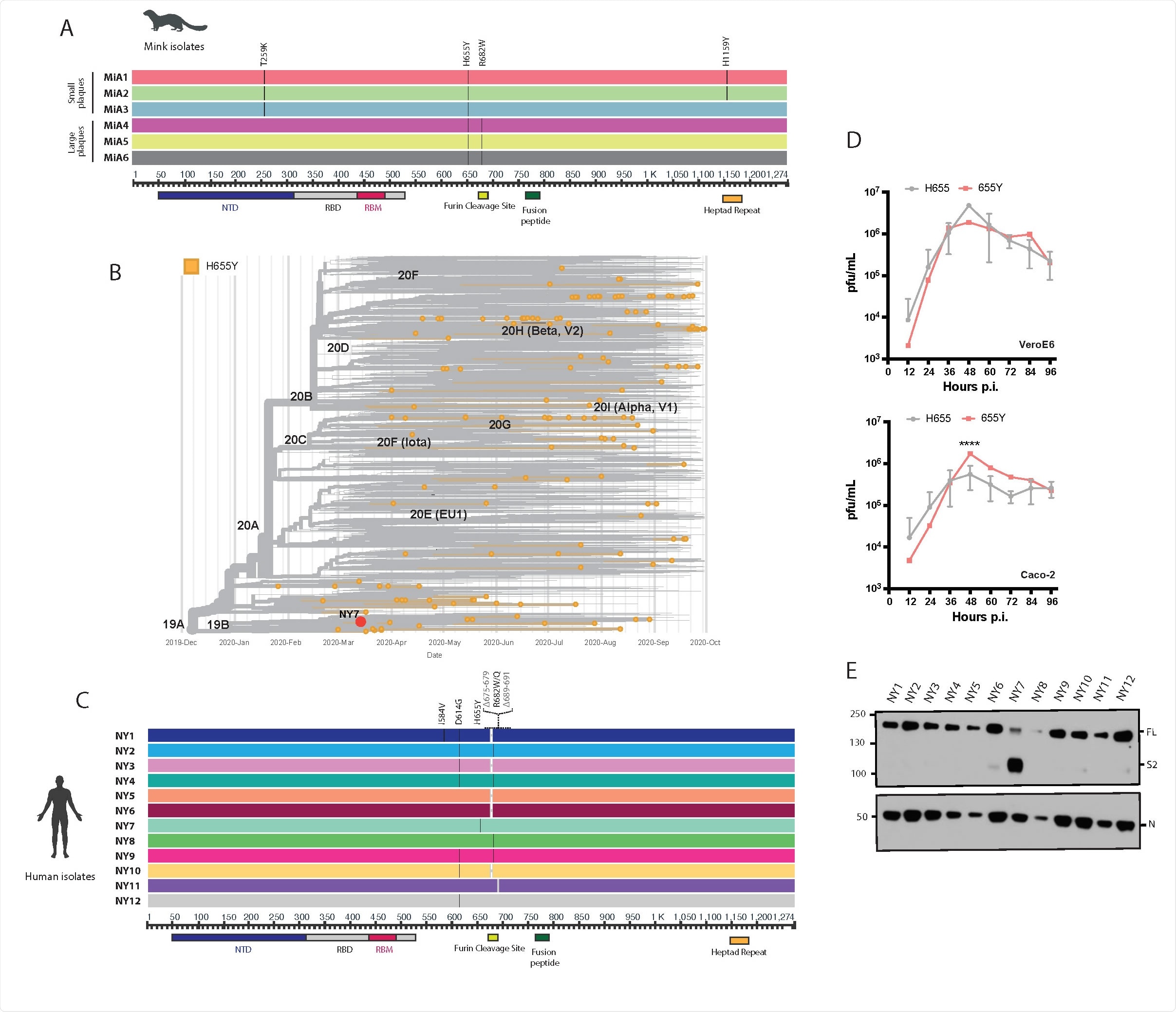
Mink and human SARS-CoV-2 variants bearing the 655Y polymorphism. A) Multiple sequence alignment of the spike (S) protein from SARS-CoV2 viruses isolated after infection in minks with WA1 isolate. Diagram shows the corresponding S amino acid substitutions mapped to the S gene. B) Time-calibrated phylogenetic analysis of the global distribution of H655Y substitution during the early SARS-CoV-2 outbreak. The phylogenetic tree was generated with Nextstrain with 7059 genomes sampled for representation of the H655Y substitution over time of worldwide data deposited in the GISAID database from December 2019 to September 2020. C) Multiple sequence alignment of the S protein from SARS-CoV2 viruses isolated from nasal swabs collected during the first pandemic wave in NY. Diagram shows the corresponding S amino acid substitutions mapped to the S gene. D) Viral growth of the NY7 containing the 655Y (red) versus its ancestors 655H (grey) in VeroE6 and Caco-2 cells. Cells were infected at an MOI of 0.01 and supernatants were titrated at the indicated hours post-infection (p.i.) and expressed as plaque forming units per milliliter (PFU). Means and SD are shown for the NY isolates containing 655H. ANOVA test was performed to compare mean differences within each group at different time points. Statistical significance was considered when p ≤ 0.05 (****, p < 0.0001). E) Western blotting of spike protein cleavage from supernatants of VeroE6 infected cells. Infections were performed at an MOI of 0.01 and supernatants were collected at 48 hours p.i. Full length (FL) spike protein (180 kDa), S2 cleaved spike (95 kDa) and Nucleocapsid (N, 50 kDa) were detected using specific antibodies. Levels of N protein were used as loading control.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
How do variants arise?
The SARS-CoV-2 variants have arisen due to several mutations in the spike protein. The first mutant that became dominant in March 2020 was S:655Y, following which many more polymorphisms arose in late 2020.
"The N501Y substitution convergently evolved in early emerging VOCs Alpha (B.1.1.7), Beta (B.1.351) and Gamma (P.1) variants and has been associated with an enhanced spike affinity for the cellular ACE2 receptor", says the team.
These mutations were located in the receptor-binding motif (RBM) of the RBD and led to the decrease in neutralizing antibody responses that were elicited by the virus.
"Similarly, the later SARS-CoV-2 Kappa (B.1.617.1) and Delta (B.1.617.2) variants have also shown a significantly reduced sensitivity to convalescent and immune sera", adds the team.
What did the study involve?
The study was carried out using the African green monkey cell line, human cell line, hamsters, and minks. The viral samples were collected from human nasopharyngeal swabs collected in March 2020 and February 2021. Variants of SARS-CoV-2 were obtained from different labs.
Next, the cell cultures, as well as hamsters and minks, were infected, and the viral titer was also calculated using the plaque assay. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to quantify the level of SARS-CoV-2 RNA present after infection.
In order to determine which mutations are mostly present at the furin cleavage site of SARS-CoV-2, mass spectrometry was used. Multiple sequence alignment was also done with the mink and human variants to identify and compare the spike mutations in them. In the case of hamsters, the frequency of variants was determined using the Oxford Nanopore Sequencing method.
What did the study find?
The results showed that the H655Y variant had a growth advantage in both human and monkey cell lines. Therefore, it can be confirmed that the S:655Y mutation alone led to enhanced growth and higher replication. Furthermore, it was observed in the hamster model as well that the efficiency of 655Y was much more than its ancestors.
Regarding the furin cleavage site, higher amounts of cleavage at this site were found in the case of alpha, delta, and kappa variants. Although the beta variant did not contain a change in the furin cleavage site, instead it contained a change in residue 701.
What did the authors conclude?
The authors concluded that the 655Y spike polymorphism was a significant cause of the SARS-CoV-2 transmission and infection.
"The selection and increasing frequency of S:655Y in the human population and following SARS-CoV-2 infection of different animal models such as cats, mice, and minks suggests this mutation is associated with an improvement of viral fitness and adaptation to diverse hosts through an increased cleavage of the spike protein", added the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources