The global death count resulting from the coronavirus disease 2019 (COVID-19) pandemic is 4.43 million. Thus, the need to maintain constant viral surveillance to detect newly emerging, possibly more pathogenic variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is paramount.
A new study details the genomic characteristics and descent of a variant of interest called B.1.621, which shows multiple amino acid substitutions within the Spike protein while discussing the impact of such variants on public health.
Study: Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Image Credit: Cristian Moga/ Shutterstock
Background
The genetic diversification of SARS-CoV-2 can increase the virus's transmissibility and enable it to escape from neutralization by antibodies generated in response to natural infection or vaccination. During the first year of the pandemic, the clock-like pattern of molecular evolution was witnessed in SARS-CoV-2.
From September 2020, substitutions started to be observed over the whole of the SARS-CoV-2 genome. Emerging variants of concern and variants of interest (VOCs and VOIs, respectively) have mutations in the spike protein. This viral glycoprotein mediates viral attachment to the host cell angiotensin-converting enzyme 2 (ACE2) receptor. Thus, the spike protein is the main target for neutralizing antibodies.
The B.1.621 lineage
The current study published in the journal Science reports on the variant of interest B.1.621 concerning its efficiency of neutralization by polyclonal antibodies, transmissibility, and binding affinity for ACE2.
The B.1 lineage surfaced in Colombia during March-April 2021, marking a change in lineage with multiple accumulated mutations. B.1.621, a member of this lineage, has the insertion 146N, along with several substitutions such as I95I,144T, and Y145S, all at the N-terminal domain; R346K, E484K, N501Y (all three at the receptor-binding domain, RBD); and P681H (this last at the S1/S2 interface).
What did the study show?
While routine monitoring was carried out in Colombia for emerging variants, by January 2021, it was strengthened to pick up imported VOCs. By the end of the first week of May, over 900 sequences had been uploaded to the GISAID database, of which the most frequent were lineage B.1, having been in circulation since the pandemic took hold.
The B.1.621 showed a rise in frequency from January 2021 to reach fifth place in frequency. The current study included all genomes from this lineage sequenced until the end of April 2021.
Phylogenetic analyses of this lineage suggested the descendence of the lineage from B.1 fairly recently, and several codons in the Spike protein were signals of positive selection. No recombination events were observed.
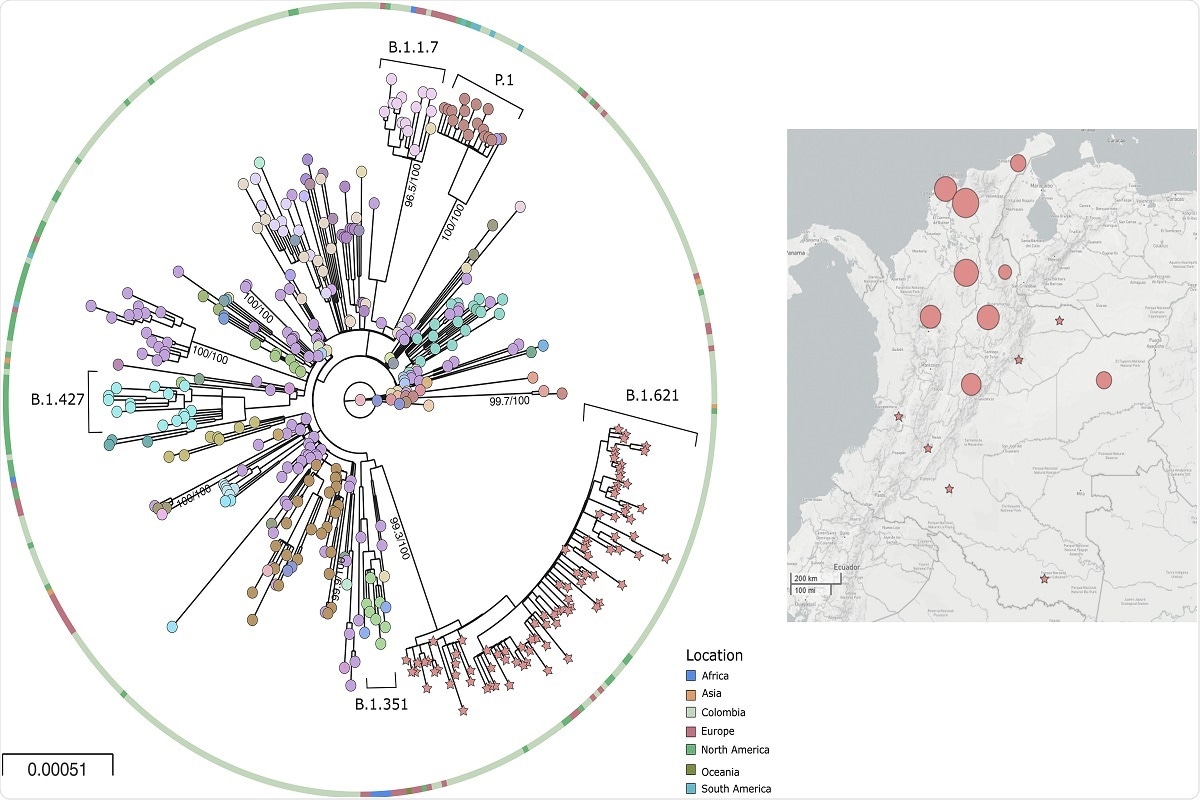
Fig. 2. Phylogeny and distribution of SARS-CoV-2 b.1.621 variant in Colombia 2a) Phylogenetic tree of the new lineage of SARS-CoV-2 emerging from B.1.621 lineage. The tree was reconstructed by maximum likelihood with the estimated GTR + F + I + G4 nucleotide substitution model for the dataset of 434 genomes. The interactive tree can be accessed in the following link: https://microreact.org/project/5CAiK3qCMaEgE4vYkKVpZW/b7113efc 2b) Map of distribution of lineages across the country.
What are the implications?
The social weariness of pandemic guidelines and the genetic characteristics of the B.1.621 strain itself led to changes in the transmissibility of this VOI. The combat of which was aided by increasing genomic surveillance for SARS-CoV-2 during the third wave of COVID-19 in the country.
Genomic sequencing is projected to cover one percent of all cases, allowing for a better picture of the real frequency of this lineage in Colombia and whether its dominance is rising or falling
The convergent substitutions seen in SARS-CoV-2 lineages worldwide are probably due to the high rate of genetic variability within the very high numbers of unexposed people globally, coupled with the use of therapeutic monoclonal antibodies that exert selection pressures on the viral genome.
Some of these common mutations are associated with relevant changes in function. E484K confers resistance to neutralization by antibodies in convalescent plasma, in the presence of N501Y and the deletion at 69/70 (del69/70). Conversely, the 145N insertion is a new mutation, and its implications are unknown.
Importantly, some of the substitutions in B.1.621 affect residues at locations targeted by the reverse transcriptase polymerase chain reaction (RT PCR), possibly leading to interference with the test results and falsely increasing the number of cases thought to be due to other VOCs.
This lineage has now been identified in Mexico, Curacao, the USA, Spain, the Netherlands, Denmark, and Germany. Further evaluation of its biological and epidemiological contributions to the pandemic is required to assess the epidemiology of the lineage.