Many studies have established that coronavirus disease 2019 (COVID-19) is more infective in men than women, and men are more at risk of severe disease. While some have suggested that behavioral differences could explain increased transmission to men, this does not explain the more morbid prognosis.
Therefore, researchers from the University of Catania have investigated potential genes and pathways that could explain this difference. Their work is published in the journal Scientific Reports.
The scientists used the freely available severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) human protein atlas program, which provides information on the tissue and cellular expression patterns of proteins that interact with SARS-CoV-2, to determine which genes to examine.
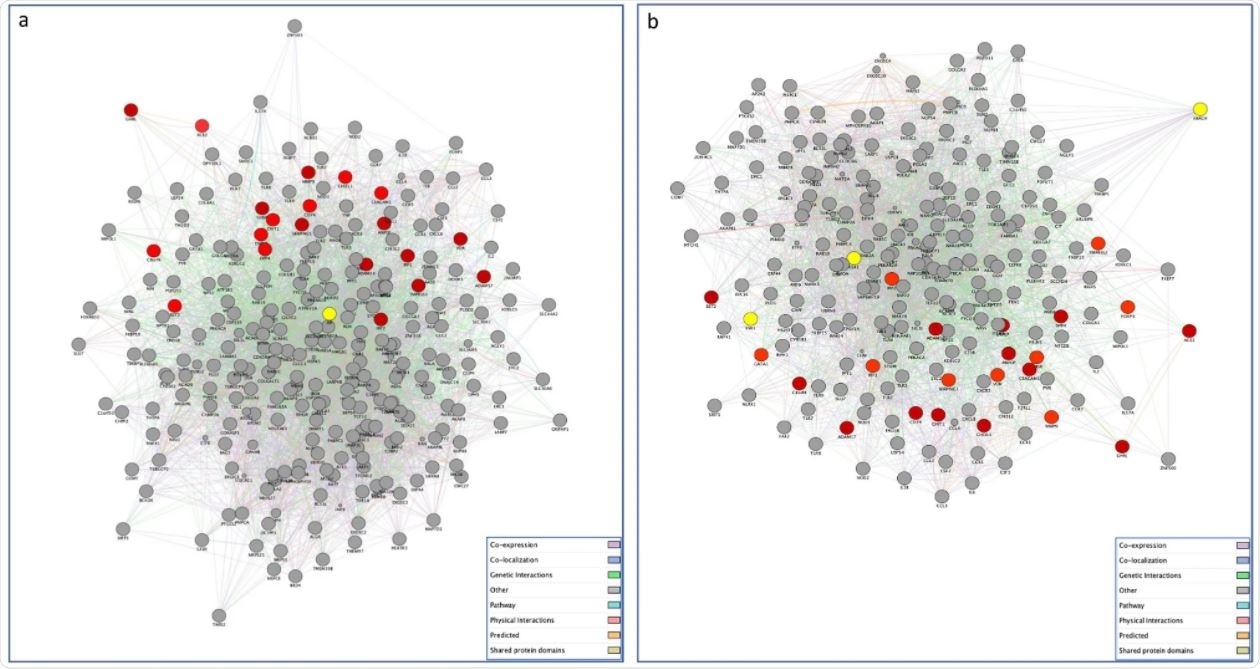
Gene networks of SARS-CoV-2 targeted human proteins expressed in testis (a) and ovary (b) and their interactions with the list of genes of our interest. The PPI network constructed by GeneMANIA shows the relationships for SARS-CoV-2 targeted human proteins expressed in testis (a) and ovary (b) and the genes of interest (nodes) connected (with edges) according to the functional association networks from the databases. Differently colored ‘edges’ indicate the type of evidence supporting each interaction: co-expression (light purple), physical interaction (pink), genetic interaction (green), shared protein domains (golden yellow), pathway (light blue), predicted (orange), and co-localization (blue).
Genes
They included genes involved with inflammatory processes such as chemokines, cytokines, metalloproteinase, bone marrow stromal antigen 2, tetherin, ANPEP, ENPEP, forkhead box P3 (FOXP3), GATA Binding Protein 1 (GATA1), High Mobility Group1 (HMGB1), Interferon Regulatory Factor (IRF1 and 2), and Serine/Cysteine Proteinase Inhibitor Clade G Member 1 (SERPING1), integrating these with the list of the HPA program and filtering on selective expression in gender-specific tissue, such as the ovary and testes. Androgen and estrogen receptors were also considered.
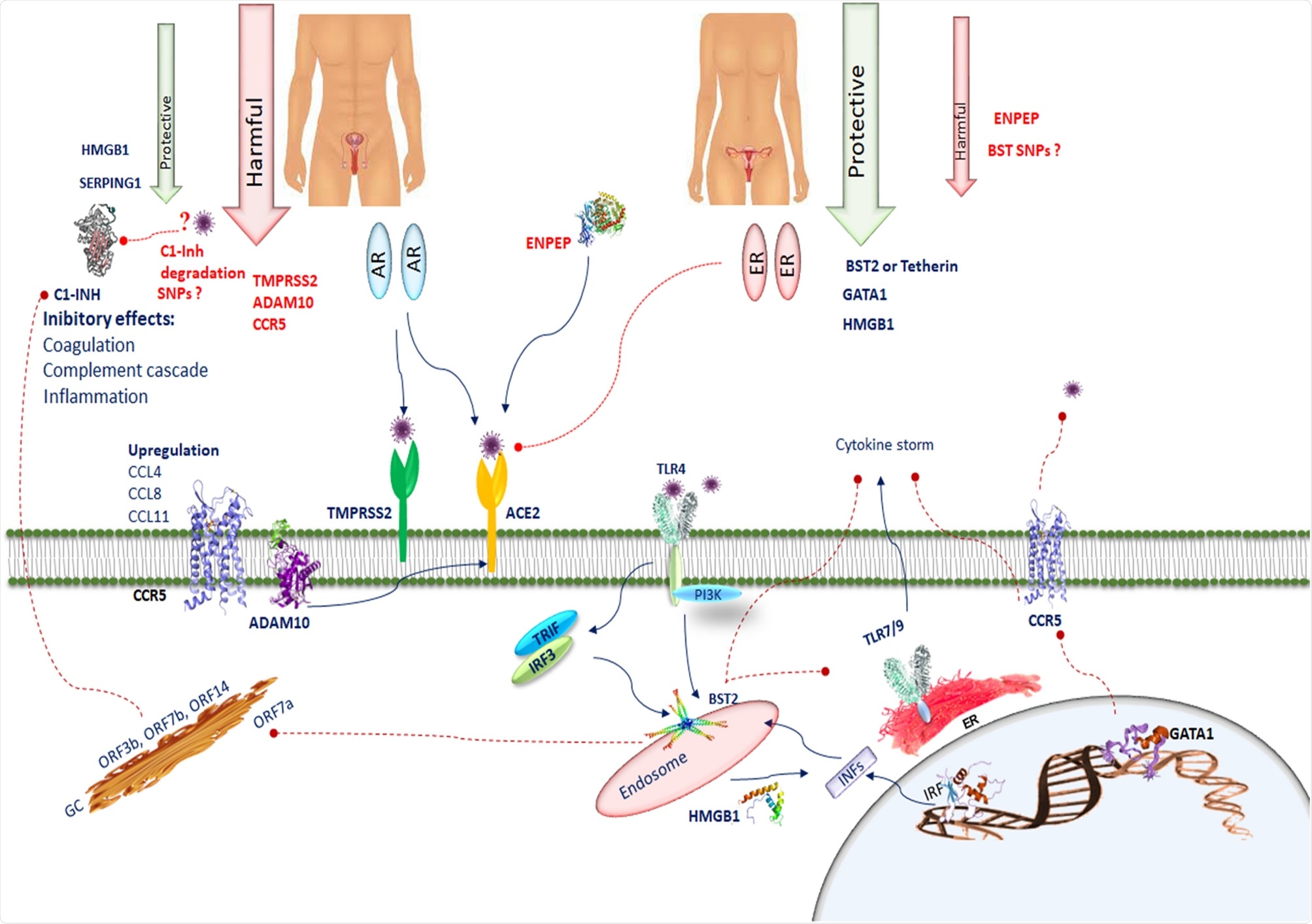
SERPING1 encodes C1INH, which suppresses complement and coagulation cascades and prevents inflammation. SERPING1 in the testis could prevent thrombotic risk. The interaction of several CoV2 proteins (ORF 3b, ORF7b, ORF14, nsp2ab, nsp13ab, nsp14ab and nsp8ab) with C1-INH can be inhibited during viral infection, leading to a predisposition to activate the complement cascade, the bradykinin pathway and the intrinsic coagulation cascade. The deletion in N-terminal region of SERPING1 by SARS-CoV-2 may block its function and increase inflammatory processes. Deteriorated SERPING1 expression caused by CoV2 interacting proteins could activate the intrinsic coagulation pathway, inducing a pro-coagulant state. CCR5 is involved in the pathology of SARS-CoV-2. In SARS-CoV-2 the chemotactic factors such as CCL4, CCL8, and CCL11 sharing CCR5 as a receptor are upregulated. ADAM10 is correlated with ACE2 cleavage regulation in human airway epithelia. BST2 reduces SARS-CoV-2 RNA replication. BST-2 is strongly induced after exposure to IFN through IRF1. BST2 expression is modulated by the TLR4/PI3K signaling pathway. Activation of TLR4 results in TRIF/IRF3-mediated positive regulation of BST-2. TLR7, TLR8 and TLR9 are predominantly localized in intracellular compartments and form the key gatekeepers in detecting and combating viral infections. In COVID-19 patients, host tetherin-mediated virion endocytosis may control TLR9 recognition to restrain immune cell responses. Several host factors such as the HMGB1 facilitate entry of self-DNA into the endosomes of pDCs, where they trigger TLR9 to induce type 1 IFN responses. GATA-1 is a potent repressor of CCR5 expression. CCR5 inhibition decreases IL-6 and SARS-CoV-2 plasma viremia. Image Credit: Spinger / Nature
GeneMANIA / Cytoscape
This list was then given to a GeneMANIA plugin in Cytoscape. GENEMANIA tests the weight from data sources based on their predicted value to re-establish the query list, generating speculation regarding the gene function and prioritizing functional genes for evaluation. Any similar genes with common characteristics are then added to the list, revealing the relationship between datasets.
Protein-protein interaction
The Cytoscape software allowed visualization of protein-protein interaction, and the researchers could then construct a gene-gene interaction network. Again, they focused on the interactions between proteins that interacted with SARS-CoV-2, inflammation-related genes, and androgen/estrogen receptors.
Gene ontology functional enrichment analysis allowed the scientists to identify overrepresented biological processes and correct using Benjami and Hochburg false discovery rate adjustment.
The researchers found that a number of SARS-CoV-2 interacting proteins expressed selectively in gonadal tissues showed significant differences between the sexes, with a total of 386 genes in the testes and 268 in the ovaries.
The GeneMania program was used to construct the aforementioned networks, with the testis-associated network consisting of 267 proteins and 5245 interactions between them.
The vast majority of these connections were co-expression, followed by physical interactions, but the scientists also found co-localization, genetic interactions, pathway interactions, shared protein domains and predicted protein interactions.
The ovary-related network showed 223 proteins and 3,726 interactions between them. These interactions followed roughly the same pattern as the male network, albeit with physical interactions taking up a more significant proportion of the total interactions.
Androgen and estrogen receptor genes
When the researchers focused their attention on androgen and estrogen receptor genes, the networks consisted of 49 proteins and 318 interactions between them (testis-related network) and 82 proteins and 634 interactions between them for the ovary-related network.
Cytoscape was then used to perform GO enrichment analysis on these networks, with most overrepresented terms associated with cell/leukocyte activation, immune response, and peptide transport. Topological network analysis identified 4 hub genes common to both networks, with radixin (RDX) showing the highest connectivity in both networks, followed by HMGB1, RAB5C and IRF2. Other high degree nodes within the testis network included SERPING1 CC chemokine receptor-5, TMPRSS2 and ADAM2. Within the ovary-related network BST2, GATA1, ENPEP, TLR4, TLR7, IRF1, and IRF2 were the high degree nodes.
The primary receptor that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) targets is angiotensin-converting enzyme 2 (ACE2). The S1 subunit of the SARS-CoV-2 spike protein contains a receptor-binding domain that binds to ACE2 in order to permit viral cell entry.
The researchers highlight high ACE2 mRNA and SARS-CoV-2 interacting protein expression levels that allow the SARS-CoV-2 virus to more easily infect cells, with several male sex hormones potentially assisting. TMPRSS2 can prime access to host cells, and activation of the androgen receptor increases TMPRSS2 levels. TMPRSS2 is also far more active in male lungs than female lungs, partially explaining the greater risk of transmission and severe disease in men. ACE2 is also regulated by the androgen receptor, which could easily facilitate the disease in the same manner.
The authors point to previous studies in mice showing the protective effect estrogen can have against lung injury. It is theorized this could be due to a reduction in ACE2 and increasing the interferon response.
The information gathered here could help explain the difference in infection and transmission between men and women and possibly help inform drug manufacturers and researchers while developing new anti-covid drugs.