The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) envelope (E) protein has a C-terminus with a PDZ binding motif (PBM) that targets PDZ (PSD-95/Dlg/ZO-1) domains. This motif is identical to the SARS-CoV PBM.
Study: Interactions of SARS-CoV-2 protein E with cell junctions and polarity PDZ-containing proteins. Image Credit: Orpheus FX / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the current study, the researchers investigated the in vitro interactions of five PDZ domains including ZO-1, LNX2, PARD3, MLLT4, and MPP5 with the full-length E protein in a PBM-dependent manner. They also studied in cellulo the role of the full-length E protein in colocalization and sequestration of PDZ domains to the Golgi compartment.
The researchers further characterized the three crystal complex structures formed by PDZ/SARS-CoV-2 E with the LNX2, MLLT4, and MPP5 proteins and focused on their specific binding modes. They also compared the binding affinities of the SARS-CoV-2 Beta variant E protein's C-terminal and SARS-CoV-2 wild-type C-terminal with the PDZ domains.
Study findings
To determine the interaction between the selected five host PDZ domains and full-length E protein of SARS-CoV-2, the researchers tagged the five PDZ domains with glutathione S-transferase (GST) and tagged the full-length E protein with green fluorescence protein (GFP). The results indicated a PBM-dependent interaction between the PDZ domains of ZO-1, LNX2, MLLT4, and MPP5 and the full-length E protein of the wild-type virus with varying binding intensities, with PARD3 being the only exception.
The researchers used a molecular replacement technique to characterize the crystal complex structure formed by the C-terminal sequence of the E protein by PBM-dependent interaction with the PDZ domains of MPP5, MLLT4, and LNX2. The PDZ domains of MPP5, MLLT4, and LNX2 bound to SARS-CoV-2-E PBM peptides with similar binding modes at positions 0 and -2.
Certainly, the N-terminal of the α2 helix is occupied by valine (Val 314), glutamine (Gln 1071), and proline (Pro 400) in PDZ domains of MPP5, MLLT4, and LNX2, respectively. The position -3 of the aspartic acid side chain forms hydrogen (H-bond) with PDZ domains of MPP5 and MLLT4; however, no interaction of proline at position -4 was found with any of the PDZ domains.
In the in cellulo study, the transfection of the SARS-CoV-2 E protein with GFP alone showed no accumulation of GFP-tagged PDZ domains. However, when GFP was co-transfected with ALFA-E, the SARS-CoV-2 E protein was involved in the relocalization and recruitment of PDZ domains from ZO1, MLLT4, MPP5, LNX2, and PARD3 into the Golgi compartment. This indicates that the SARS-CoV-2-E protein can potentially bind to PDZ domains in a PBM-dependent manner.
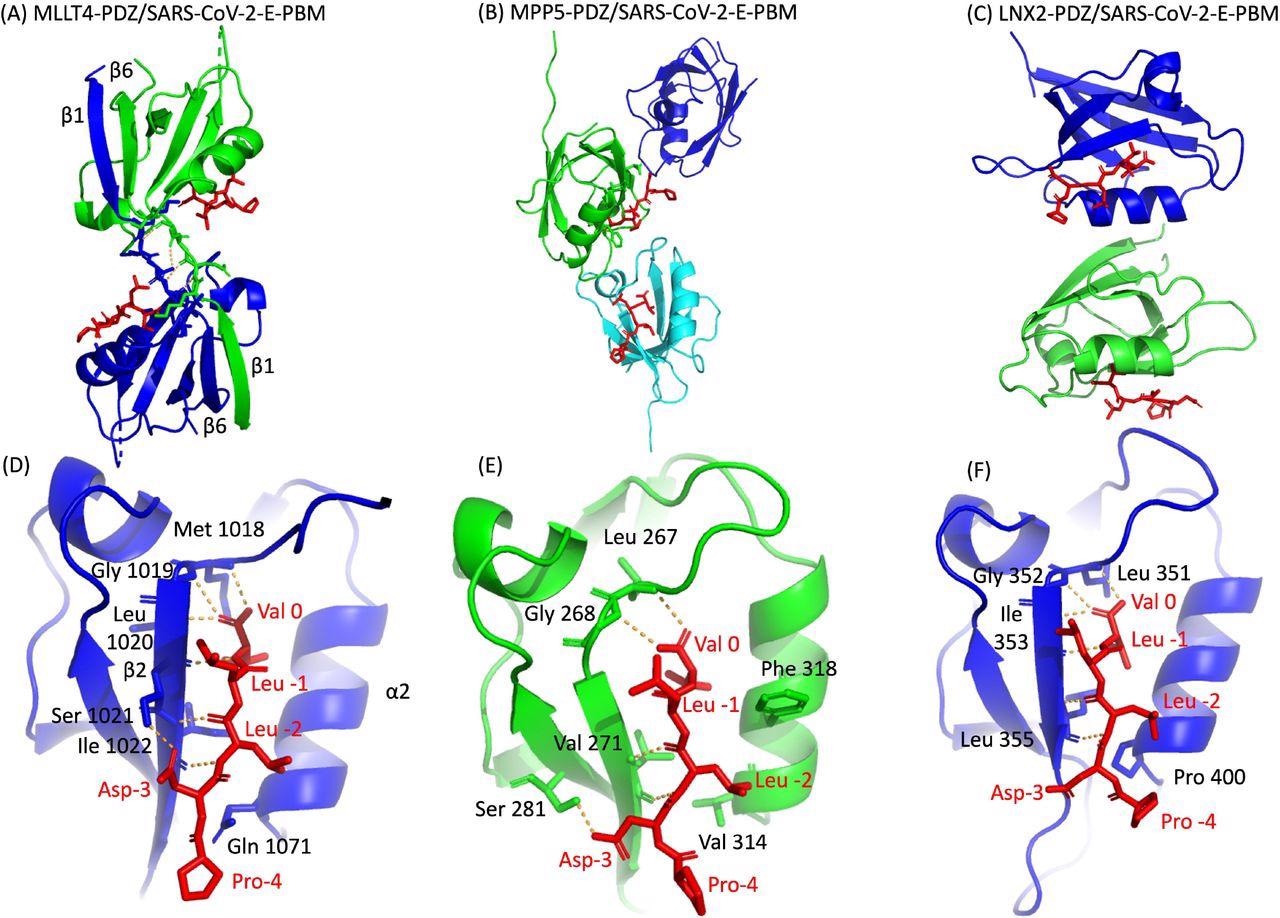 X-ray structures of PDZ domains of MLLT4, MPP5 and LNX2 bound to the SARS-CoV-2 protein E PBM. (A)(B)(C) The asymmetric unit of the PDZ domains of MLLT4, MPP5 and LNX2 respectively, bound to the SARS-CoV-2 protein E PBM shown as red sticks. (A) Selected interchain polar contacts related to the swapped dimer between the fragments Lys 1014-Gly 1017 of each chain are shown in orange and the associated residues are shown as sticks. (D)(E)(F) Detailed views of the PDZ domains bound to SARS-CoV-2 protein E PBM. Important residues are labeled and shown as sticks. Intermolecular H-bonds and polar contacts are reported as orange dashed lines.
X-ray structures of PDZ domains of MLLT4, MPP5 and LNX2 bound to the SARS-CoV-2 protein E PBM. (A)(B)(C) The asymmetric unit of the PDZ domains of MLLT4, MPP5 and LNX2 respectively, bound to the SARS-CoV-2 protein E PBM shown as red sticks. (A) Selected interchain polar contacts related to the swapped dimer between the fragments Lys 1014-Gly 1017 of each chain are shown in orange and the associated residues are shown as sticks. (D)(E)(F) Detailed views of the PDZ domains bound to SARS-CoV-2 protein E PBM. Important residues are labeled and shown as sticks. Intermolecular H-bonds and polar contacts are reported as orange dashed lines.
Binding affinities of PDZ domains
The current study determined the binding affinity of the original SARS-CoV-2 E protein C-terminal PBM peptide and the Beta variant P71L PBM peptide for the PDZ domains of ZO1, MLLT4, MPP5, LNX2, and PARD3 using the microscale thermophoresis technique.
For the original SARS-CoV-2 E peptide, the PDZ of MPP5 and the PDZ2 of ZO-1 displayed the best affinities with Kd values of 30 micromolar (μM) and 15 μM, respectively. The MLLT4-PDZ and PARD3-PDZ3 affinities were lower, with Kd values of 569 µM and 341 µM, respectively.
For the Beta variant P71L mutation, the affinities for the PDZ domains of MLLT4 and PARD3 were not detectable in the tested concentration range, thereby suggesting that they might not react with PDZ domains of MLLT4 and PARD3 or else the binding affinities were beyond the tested concentration range. The mutation P71L had no significant impact on the binding affinity of MPP5 but significantly reduced the affinity for ZO-1-PDZ2 with a Kd of 133 µM. The affinity of LNX2-PDZ2 improved from 289 µM to 47 µM in mutant P71L.
Overall, the results show that the four PDZ domains MPP5, ZO-1, MLLT4, and PARD3 exhibit different binding affinities for P71L. This suggests a differential specificity profile of the original C-terminal SARS-CoV-2 E protein PBM and the Beta C-terminal SARS-CoV-2 E protein PBM P71L for the PDZ domain.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zhu, Y., Alvarez, F., Wolff, N., et al. (2021). Interactions of SARS-CoV-2 protein E with cell junctions and polarity PDZ-containing proteins. bioRxiv. doi:10.1101/2021.12.04.471219. https://www.biorxiv.org/content/10.1101/2021.12.04.471219v1.
- Peer reviewed and published scientific report.
Zhu, Yanlei, Flavio Alvarez, Nicolas Wolff, Ariel Mechaly, Sébastien Brûlé, Benoit Neitthoffer, Sandrine Etienne-Manneville, Ahmed Haouz, Batiste Boëda, and Célia Caillet-Saguy. 2022. “Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E with Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.” Frontiers in Microbiology 13 (February). https://doi.org/10.3389/fmicb.2022.829094. https://www.frontiersin.org/articles/10.3389/fmicb.2022.829094.