Widespread vaccination efforts against the coronavirus disease 2019 (COVID-19) have successfully prevented innumerable severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections, hospitalizations, and deaths. Despite these efforts, the emergence of immune-evading SARS-CoV-2 variants has significantly reduced the efficacy of current COVID-19 vaccines.
Given the ongoing evolution of SARS-CoV-2, researchers are working to determine whether current COVID-19 vaccines should be updated to match circulating variations antigenically. These efforts would closely resemble vaccine development for seasonal influenza viruses, which require periodic vaccine updates due to antigenic drift.
Study: Mapping the antigenic diversification of SARS-CoV-2. Image Credit: CROCOTHERY / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In a recent study published on the medRxiv* preprint server, researchers investigate SARS-CoV-2 antigenic drift by evaluating the neutralizing activity of a unique set of sera from patients infected with all five SARS-CoV-2 variants of concern (VOCs).
The authors collected and analyzed serum samples from 51 COVID-19 patients who had a primary SARS-CoV-2 infection verified by reverse-transcriptase polymerase chain reaction (PCR) assay but had not received COVID-19 vaccines. Three to nine weeks following the onset of symptoms, blood was collected to determine the peak of the antibody response.
Of the 51 subjects included in the current study, 31 had sequence-confirmed VOC infection. More specifically, ten, seven, four, and ten participants were positive for the Alpha, Beta, Gamma, and Delta variants, respectively. As the remaining 20 samples were acquired before the emergence of any VOC in the Netherlands, these individuals were believed to be infected with the ancestral strain.
The authors evaluated the neutralizing power of the convalescent sera samples against the ancestral SARS-CoV-2 strain, as well as the Alpha, Beta, Gamma, Delta, and Omicron variants using a lentiviral-based pseudovirus neutralization assay. As expected, the highest neutralization titers were often recorded against the homologous virus.
Study findings
Only subjects infected with the Beta variant had significantly stronger cross-neutralization titers against the Gamma variant as compared to homologous neutralization. When non-hospitalized patients were compared, those infected with the Alpha variant demonstrated the highest level of homologous neutralization, followed by those infected with the Gamma strain, ancestor strain, and Delta variant. By contrast, none of the participants infected with the Beta variant demonstrated significant homologous neutralization.
The ability of SARS-CoV-2 VOCs to generate cross-neutralizing antibodies varied significantly. To this end, individuals infected with the Alpha variant elicited the largest response, followed by those infected with the original, Gamma, and Delta strains; however, there was considerable variation within each group. No patient infected with the Beta variant showed significant cross-neutralization activity.
All groups experienced significant decreases in neutralizing activity against the Omicron variant. Omicron neutralization fell below the detection limit in 21 of the persons examined.
When all patients were considered, the fold reduction in Omicron neutralization against homologous neutralization was nine-fold, ten-fold for patients infected with ancestral strains, 56-fold for Alpha, six-fold for Gamma, and five-fold for Delta-infected individuals.
The authors created an antigenic map of SARS-CoV-2 using neutralization data. It is clear from this map that homologous neutralization is dominating.
Furthermore, this map demonstrates that the ancestral and Alpha strains of SARS-CoV-2 are closely related, whereas the Beta, Gamma, and Delta variants are all within two antigenic units of the ancestor virus. In the case of influenza viruses, antigenic distances of less than three antigenic units are considered comparable.
These antigenic clusters are related to one another through analogy. The fact that the Omicron variant is the first extensively circulating new SARS-CoV-2 variant is implied by the distance between this antigenic cluster and Omicron.
Concerns about the comparability of pseudovirus neutralization titers to hemagglutination inhibition assay titers used to define influenza viruses are raised. The difference between Omicron and other SARS-CoV-2 variants, including the progenitor strain, is significant.
To create a second antigenic map, the authors used neutralization data from 122 COVID-19 naive vaccinees who received two Moderna or AstraZeneca vaccinations based on the ancestral S sequence. This map correlated well with the infectee map, thus confirming that Omicron is a separate antigenic variation from vaccine viruses.
Serum from different vaccine recipients overlaps, although mRNA-1273 vaccinees have a slight bias towards Omicron. This indicates slight changes in antigen stimulation among the vaccine formulations evaluated here.
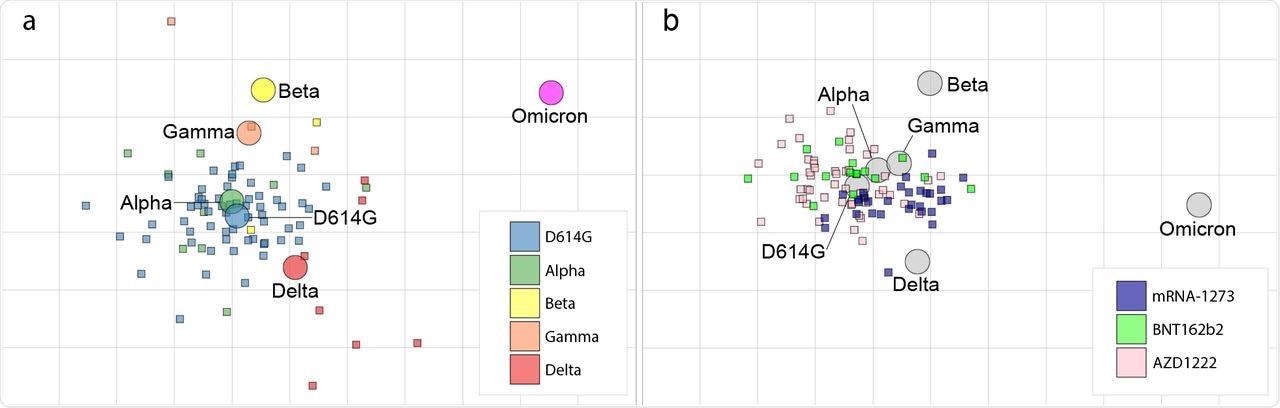
SARS-CoV-2 antigenic cartography.
Implications
As is the situation with seasonal influenza viruses, the possibility of SARS-CoV-2 becoming an endemic virus with repeated outbreaks necessitates the need for antigenic drift surveillance and the administration of new immunizations on an annual basis, particularly for those at risk for severe COVID-19. Antigenic mapping initiatives, such as those described in this study, can provide valuable information for future vaccine development.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Van der Straten, K., Guerra, D., van Gils, M. J., et al. (2021). Mapping the antigenic diversification of SARS-CoV-2. medRxiv. doi:10.1101/2022.01.03.21268582. https://www.medrxiv.org/content/10.1101/2022.01.03.21268582v1.
- Peer reviewed and published scientific report.
Straten, Karlijn van der, Denise Guerra, Marit J. van Gils, Ilja Bontjer, Tom G. Caniels, Hugo D. G. van Willigen, Elke Wynberg, et al. 2022. “Antigenic Cartography Using Sera from Sequence-Confirmed SARS-CoV-2 Variants of Concern Infections Reveals Antigenic Divergence of Omicron.” Immunity, August. https://doi.org/10.1016/j.immuni.2022.07.018. https://www.cell.com/immunity/fulltext/S1074-7613(22)00353-3.