
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
With the ongoing COVID-19 pandemic causing devastation on a global scale, scientists and public health systems alike have been working together to address the challenges the pandemic entails and develop policies to control it.
Since the pandemic began, scientific research has grown exponentially at an unparalleled pace, from exploring and testing therapeutic drugs to developing vaccines against SARS-CoV-2. Data suggests that over 52,000 peer-reviewed articles were published during the first year of the COVID-19 crisis, as compared to around 1,000 during the initial 12 months of the SARS outbreak in 2002.
The staggering magnitude of research data on COVID-19 and SARS-CoV-2, which continues to expand, requires a combined database to house the research data from across various available repositories in a standardized, searchable, interpretable, and easy-to-access interface.
The pandemic has led to the creation of several databases and for instance, numerous websites report COVID-19 cases across different geographical regions that are mostly contributed by volunteers.
LitCovid is a hub of the COVID-19 literature, while the data on clinical trials are stored at the National Clinical Trials (NCT) registry. Therefore, a common library that provides access to COVID-19 resources assembled from various sources is required to aid scientific research.
In the present paper, the authors describe the development of outbreak.info. This website hosts COVID-19 research data created by collecting metadata from 14 repositories and combining COVID-19 resources from hundreds of sources scattered over the internet and yet remain disparate.
The database hosts data resources from over 200,000 publications, clinical trials, and other related datasets. The collected resources were standardized by developing schema, prioritizing five classes of COVID-19 research data – publications, datasets, clinical trials, analysis, and protocols.
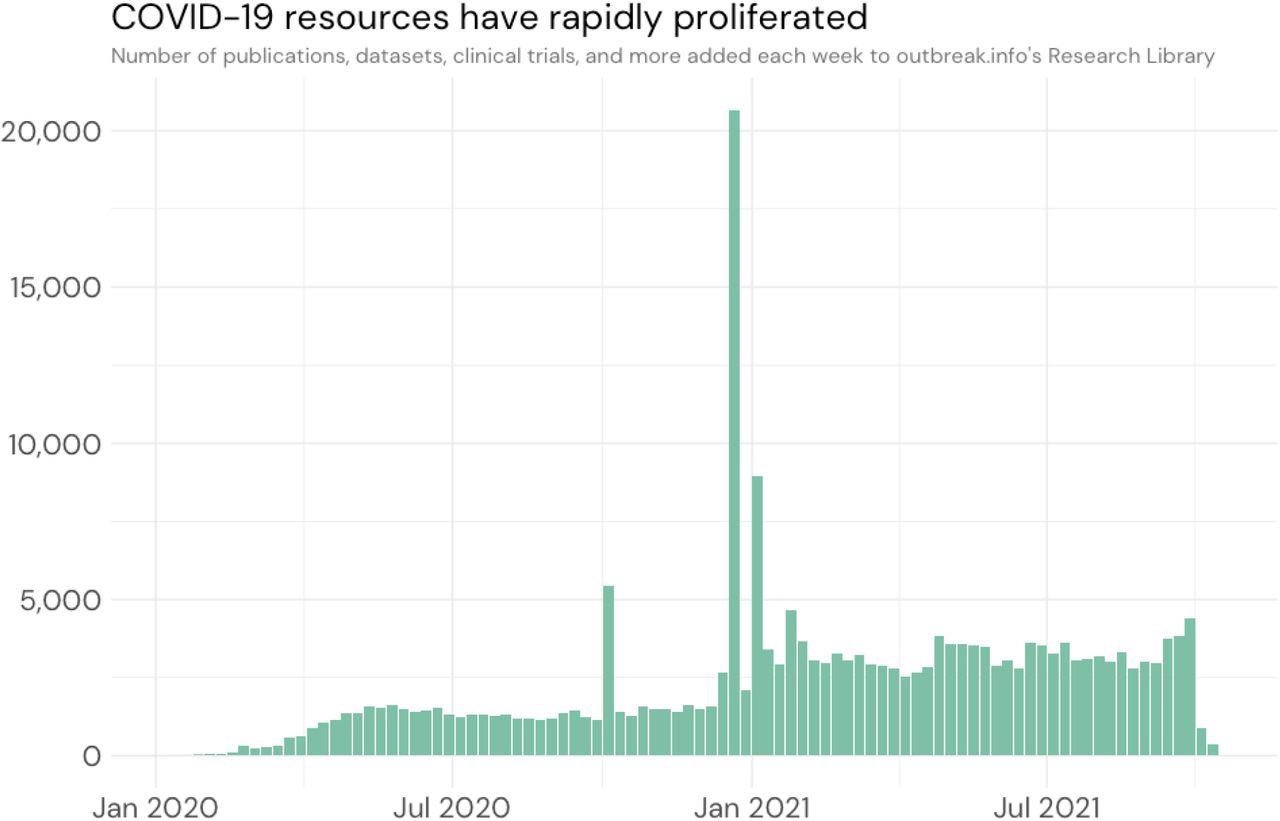
Number of resources in outbreak.info as a function of date.
Metadata is ingested into the website in two ways. For example, the first method uses the BioThings software development kit (SDK) data plugins, and the second method allows submissions via an online form. A nested list of thematic or topic-based categories was developed based on the initial list from LitCovid, which resulted in a list with 11 broad categories and 24 specific child categories. Epidemiological data was ingested from John Hopkins University (JHU) and the New York Times (NYT), and the genomics data was integrated from the GISAID database.
Findings
After developing the schema, the researchers created data plugins or parsers to import metadata from 14 repositories and ingest it into outbreak.info. These parsers auto-update daily to maintain updated information. The most extensive data class was publications collected from LitCovid and the preprint servers, bioRxiv and medRxiv. The clinical trial data from the NCT and World Health Organization (WHO) formed the second largest library. The "protocols" class compiled data from two resources - Protocols.io and NCT protocols, while the datasets library sourced its information from Zenodo, protein data bank (PDB), Figshare, and Harvard datasets.
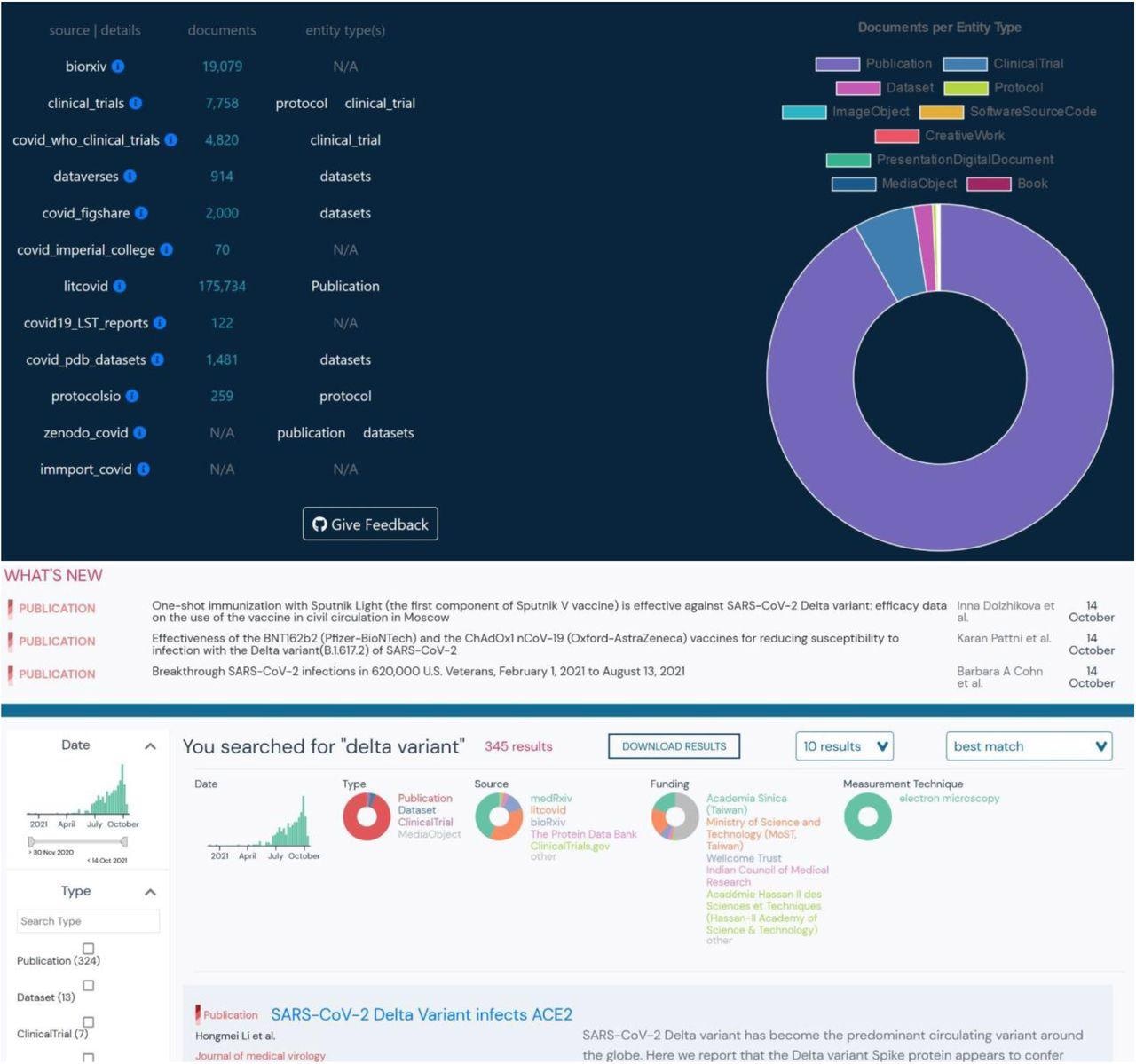
A. Distribution of resources by resource type and source. B. Heterogeneous and filterable resources (ie-publications, clinical trials, datasets, etc.) resulting from a single search of the phrase “Delta Variant”
Data available at the Imperial College of London (ICL) were imported to fill the "Analysis" library class. The database has been developed with a feature to allow submissions from the "volunteers" or the community. Other features include creative and interactive visualization of epidemiological data imported from JHU and NYU, although many other sources compile information on epidemiology from JHU, the interface on outbreak.info is built to support research.
Conclusions
The authors of the present work have created a database to access resources of COVID-19 and SARS-CoV-2 easily. The massive expansion of research and epidemiological data necessitates a shared library that houses information from many sources in an easy, searchable, standardized, and interpretable interface. This has been achieved by creating outbreak.info, a feature-rich website that allows contributions from the community. Furthermore, the integration of data compiled from various repositories into a single database allows quick exploration and retrieval of COVID-19 resources irrespective of their source.
In summary, the authors created a website that essentially comprises three components: 1) outbreak.info contains a searchable interface, 2) a tool to explore epidemiology data and spatiotemporal trends, and 3) surveillance reports on SARS-CoV-2 variants and mutants. The website is also integrated with public application programming interfaces or APIs to allow access to resource data.
What is Outbreak.info? The Open-Source Hub of COVID-19 Data & Research

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tsueng, Ginger, Julia Mullen, Manar Alkuzweny, Marco Alvarado Cano, Benjamin Rush, Emily Haag, Outbreak Curators, et al. "Outbreak.Info: A Standardized, Searchable Platform to Discover and Explore COVID-19 Resources and Data." bioRxiv, January 21, 2022. DOI: https://doi.org/10.1101/2022.01.20.477133, https://www.biorxiv.org/content/10.1101/2022.01.20.477133v1
- Peer reviewed and published scientific report.
Tsueng, Ginger, Julia L. Mullen, Manar Alkuzweny, Marco Cano, Benjamin Rush, Emily Haag, Jason Lin, et al. 2023. “Outbreak.info Research Library: A Standardized, Searchable Platform to Discover and Explore COVID-19 Resources.” Nature Methods, February. https://doi.org/10.1038/s41592-023-01770-w. https://www.nature.com/articles/s41592-023-01770-w.