
 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
With a mutation rate of approximately 1.1 × 10-3 substitutions/site/year, several SARS-CoV-2 VOCs have emerged that are highly infectious and transmissible. While the Alpha and Delta variants resulted in the second and third coronavirus disease 2019 (COVID-19) pandemic waves, respectively, most recently, in November 2021, the SARS-CoV-2 variant Omicron variant emerged in South Africa and subsequently led to the fourth pandemic wave around the world.
The SARS-CoV-2 D614G strain observed in May 2020 consists of a single mutation and was highly infectious. Although D614G infection resulted in a high viral load in the upper respiratory tract, its infection did not significantly alter disease severity as compared to the ancestral SARS-CoV-2 Wuhan-1 strain.
The first SARS-CoV-2 VOC to be identified was the Alpha variant, which also had the D614G mutation and a cleavage site mutation P681H. Comparatively, the SARS-CoV-2 Delta variant also had D614G and P681R mutations in the spike (S) domain.
The recently identified Omicron VOC harbors the D614G and P681H mutations, as well as a mutated S proline 681 to histidine instead of arginine, which was also observed in the Delta variant.
There remains a limited amount of studies that have evaluated the impact of D614G and P681H/R mutations on the traits of VOCs. However, it is crucial to understand the mechanisms behind the entry and fusion of emerging VOCs, especially as they provide immune escape ability to these VOCs and reduce the neutralization efficacy of current vaccines.
About the study
In the present study, researchers used the full-length wild-type ancestral Wuhan-1 S protein and subjected it to site-directed mutagenesis. To this end, the researchers synthesized the D614G, Alpha, Delta, Omicron, single P681H mutant, and D614G+P681R double mutant pseudoviruses.
The researchers prepared pseudoviruses in 293T cells containing furin-like proteases. As compared to the Delta pseudovirus, Alpha and Omicron pseudoviruses showed slower S cleavage processing in the presence of furin. All mutants were well-expressed in BHK-21 transfected cells.
S protein expression of all the variant and mutant pseudoviruses on the cell surface, as compared to wild type S, was assessed using flow cytometry in HEK293T mammalian cells. While analyzing the binding of surface-expressed S protein to soluble human angiotensin-converting enzyme 2 (hACE2) by flow cytometry, the authors noted that the Delta, Omicron, and D614G+P681R S mutants showed a two-fold increase in binding to soluble hACE2.
The researchers also analyzed the cleavage of S protein in pseudovirions at 24 and 48 hours post-transfection. Next, they analyzed concentrated pseudovirions using Western blot.
Except for Alpha pseudovirus S, the S proteins of all other variants were cleaved at 24 hours post-transfection. Comparatively, for the Omicron pseudovirus, S cleavage was observable at 48 hours post-transfection.
Study findings
The Delta variant showed enhanced virus entry in hACE2 and transmembrane protease, serine 2 (TMPRSS2) overexpressed cells. On the other hand, the double mutant D614G+P681R showed a marked increase in virus entry through both hACE2 and TMPRSS2 receptors, similar to Delta pseudovirus.
The synthetic double mutant D614G+P681R also showed high infectivity titer in both hACE2 and TMPRSS2 overexpressed cells. This indicates the potential role of these two mutations in hACE2 and TMPRSS2 mediated virus entry.
The Omicron pseudovirus showed hACE2-dependent virus entry, low fusion, syncytial formation, and reduced infectivity titers. Interestingly, the marked reduction in fusion capacity of Omicron pseudoviruses was due to the presence of P681H mutation in the Omicron S protein. Further, all variants showed a significant increase in infectivity titers in 293T-hACE2 cells.
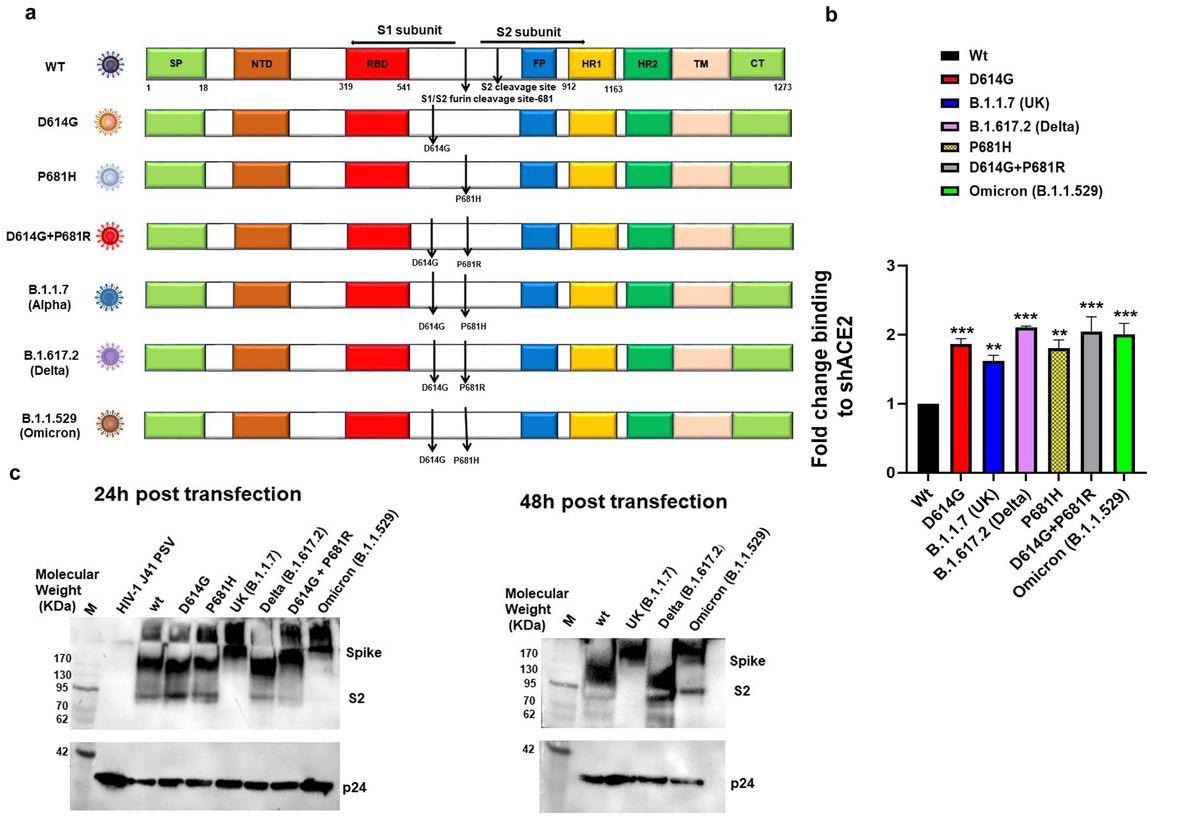
Expression and cleavage of SARS-CoV-2 variants and synthetic mutants: a. Schematic representation of ancestral, Alpha, Delta and Omicron spike protein with D614G and P681R/H mutations and synthetic mutants, the signal peptide (green box), S1-N terminal domain NTD (brown box), Receptor binding domain (Red box), fusion peptide (blue box),Heptad repeat 1(yellow box), Heptad repeat 2(dark green box), Transmembrane domain TM (Cream box), Cytoplasmic tail CT (green box), S1/S2 and S2′ cleavage site, presence of D614G, P681H/R mutation b. Surface expressed spike protein binding to soluble-hACE2 was analyzed by expressing the spike of variants and mutants in HEK 293T cells and 36 h post-transfection incubated with soluble-hACE2 and analyzed by flow cytometry. Each value represents a single mean value of two repeated experiments. Statistical significance was determined using the one-way ANOVA test (p < 0.05), where *** and ** p < 0.05 is significant. c. Detection of spike protein cleavage in different pseudoviruses at 24h and 48h post-transfection by Western blot analysis, p24 was kept as loading control.
Further assessment of SARS-CoV-2 variant S proteins showed that the P681R mutation allowed increased cell-to-cell fusion in the presence of exogenous trypsin in the Delta and double D614G+P681R mutant S proteins. This phenomenon was not observed with the Omicron S protein, which has the P681H mutation and single P681H mutant. These findings suggest that the P681H mutation in the Omicron S cleavage site down-regulated the cleavability of S protein by furin or trypsin-like proteases, thus restricting localized replication of Omicron.
The authors also noted that Omicron pseudovirus titers increased in hACE2 cells and virus entry was reduced to more than 50% in the presence of cathepsin L inhibitor E64d. However, in the presence of TMPRSS2 inhibitor camostat mesylate, there was a reduction of about 34% in the entry to host cells. These findings suggested that Omicron preferentially entered host cells through ACE2-mediated endocytosis.
Conclusions
Overall, the SARS-CoV-2 Delta and Omicron VOCs demonstrated the evolution of both virulent and moderately virulent strains for better fitness for survival. In the Delta variant, the presence of the D614G and P681R mutations was crucial for virulence and pathogenesis.
Comparatively, in the Omicron variant, the P681H mutation resulted in slow processing of S cleavage by furin or trypsin-like proteases and allowed ACE2-dependent entry. This subsequently restricted Omicron replication mainly to the upper respiratory tract. The single synthetic mutant P681H also closely resembled Omicron in its behavior.
Developing an understanding of the SARS-CoV-2 entry pathway will allow for the appropriate selection of cell lines for in vitro screening of drugs and vaccine candidates.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Khatri, R., Siddqui, G., Sadhu, S., et al. (2022). SARS-CoV-2 variants’-Alpha, Delta, and Omicron D614G and P681R/H mutations impact virus entry, fusion, and infectivity. Research Square. doi:10.21203/rs.3.rs-1310197/v1.