Throughout each stage of its life cycle, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interacts with host membranes. Moreover, SARS-CoV-2 penetrates the cell by crossing the plasma membrane, thus replicating inside host-derived membrane compartments. SARS-CoV-2 subsequently obtains its envelope from the host and exits the cell through the Golgi apparatus and lysosome.
For viruses to fulfill their structural, metabolic, and trafficking requirements, they must rely on host pathways; however, viruses often need to modify these pathways to be effective. SARS-CoV-2, for example, re-engineers the host internal membranes into double-membraned vesicles (DMVs) and convoluted membrane (CM) areas to enable replication.
Study: A global lipid map reveals host dependency factors conserved across 2 SARS-CoV-2 variants. Image Credit: Kateryna Kon / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
SARS-CoV-2 and lipids
Several previous studies have found that SARS-CoV-2 infection involves the modulation of host lipids as a core characteristic. Several lipids and lipid-associated proteins, such as very-low-density lipoproteins (VLDL) and high-density lipoproteins (HDL), steroid hormones, and various apolipoproteins have been identified as biomarkers of infection.
Furthermore, elevated triacylglycerol (TAG) and polyunsaturated free fatty acids are suspected of being involved as markers of severe disease outcomes. Obesity, diabetes, and hypertension have also been identified as important risk factors in people who acquire severe COVID-19.
In a recent study posted to the preprint server bioRxiv*, SARS-CoV-2 appears to alter host lipid biosynthesis by relying on certain human metabolic pathways to survive and multiply effectively. To come to this conclusion, the authors conducted an intricate lipid study of both infected cells and cells ectopically expressing individual SARS-CoV-2 proteins.
Assessing changes in lipoprotein levels
The researchers first conducted global lipidomic profiling of HEK293T cells that overexpress the angiotensin-converting enzyme 2 (ACE2) receptor, which is primarily used by the SARS-CoV-2 spike protein to enter host cells. These cells were then infected with either SARS-CoV-2 or a control virus for 24 hours, which was followed by an analysis of cellular lipids through liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS).
Taken together, a total of 514 lipids were identified in HEK293T cells. Of these lipids, 79.6% differed between the SARS-CoV-2 and mock-infected cells, the levels of which changed between2- and 64-fold following infection.
The researchers then sought to understand how lipid composition was altered following exposure to SARS-CoV-2. To this end, the most significant changes were observed in both glycerolipids and phospholipids, with TAG and cardiolipin (CL) exhibiting the greatest changes, with changes in the TAG species being dependent upon their fatty acid composition. More specifically, TAG species with polyunsaturated fatty acid (PUFA) chains were found to be eight times more abundant than saturated or monounsaturated species in SARS-CoV-2 infected cells.
When examining the changes that occurred in phospholipids following SARS-CoV-2 infection, several saturated phospholipids declined, of which included phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and phosphatidylinositol (PI). Notably, PC and PG were reduced by 1.5- and 1.7-fold, respectively. Comparatively, several polyunsaturated species increased including plasmalogen-linked phosphatidylcholine (P-PC), which increased by 2.7-fold.
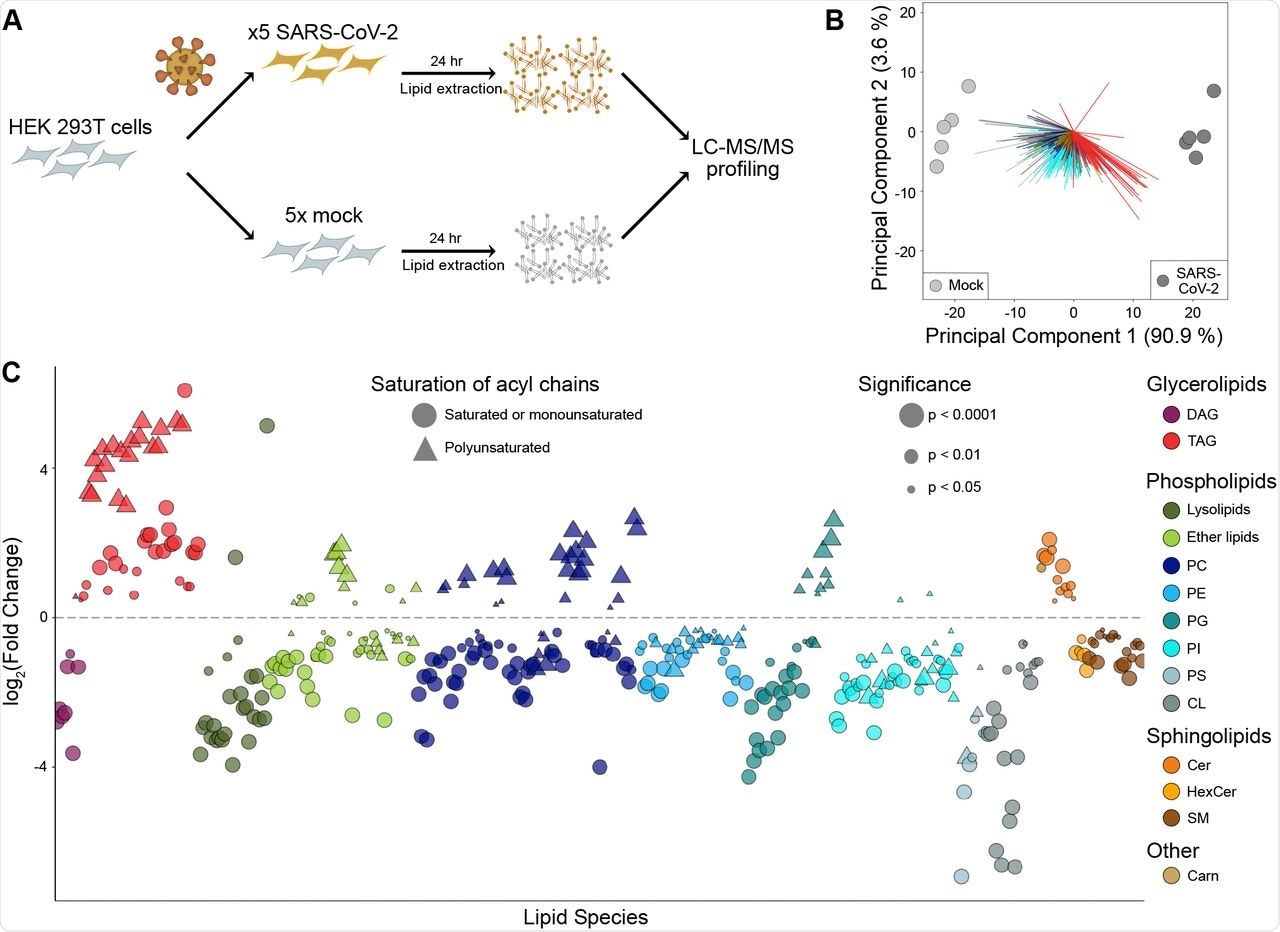 SARS-CoV-2 alters the lipid composition of its host cells.
SARS-CoV-2 alters the lipid composition of its host cells.
Lipidomic analysis of SARS-CoV-2-infected cells
Untargeted lipidomic profiling of cells transfected with each SARS-CoV-2 protein was then performed, aside from nsp3, nsp14, nsp15, nsp16, and orf3b due to transfection efficiency challenges. After 48 hours, total cellular lipids were extracted from these proteins and subsequently analyzed by LC-ESI-MS/MS.
Taken together, 396 unique lipids were identified from the SARS-CoV-2 proteins, which consisted of glycerolipids, phospholipids, sphingolipids, and acyl-carnite lipids. Of these lipids, 80% were significantly changed in at least one transfection with a given SARS-CoV-2 protein.
The most common lipids that increased following transfection with a viral protein included PIs, diacylglycerols (DAGs), and ether-linked lipids, particularly vinyl-ether phosphatidylcholines (O-PC), which were elevated in 75%, 43%, and between 26% and 43% of transfections, respectively. Comparatively, the lipids that were decreased in the SARS-CoV-2 protein transfections included Lyso-PC, CL, and TAGs, which were reduced in 75%, 43%, and 50% of transfections, respectively.
A closer look at SARS-CoV-2-induced lipid changes
After acquiring the aforementioned data, the researchers looked to better understand how each SARS-CoV-2 protein affects lipid remodeling during infection. TAG, for example, exhibited an increase in remodeling by SARS-CoV-2 orf6, nsp5, orf9c, orf3a, and orf7a proteins, whereas polyunsaturated PC was impacted by orf6, orf9c, orf9b, and E.
Taken together, the findings from these experiments point to TAG as the most significantly and substantially increased lipid following SARS-CoV-2 infection. Notably, other viruses including hepatitis C virus (HCV) and rotaviruses both cause lipid droplets (LDs) to accumulate during infection, which are the cellular reservoirs for TAGs. The researchers observed a similar phenomenon to occur following infection with SARS-CoV-2, wherein LDs increased in each infected cell in a time-dependent manner.
Implications
By thoroughly examining the effect of individual viral proteins on host lipids, the researchers were able to reveal a complex network of many different SARS-CoV-2 proteins that play a role in different aspects of host lipid remodeling. Thus, the use of glycerolipid production inhibitors to prevent viral replication highlights the value of both of the lipidomic datasets in understanding COVID-19 pathophysiology and their potential as anti-SARS-CoV-2 treatment options.
Given the rapidly spreading and mutating nature of SARS-CoV-2, it is vital for scientists to understand the underlying biology of its life cycle to assist in the development of prophylactic and therapeutic options for COVID-10.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Farley, S. E., Kyle, J. E., Leier, H. C., et al. (2022). A global lipid map reveals host dependency factors conserved across 2 SARS-CoV-2 variants. bioRxiv. doi:10.1101/2022.02.14.480430. https://www.biorxiv.org/content/10.1101/2022.02.14.480430v1
- Peer reviewed and published scientific report.
Farley, Scotland E., Jennifer E. Kyle, Hans C. Leier, Lisa M. Bramer, Jules B. Weinstein, Timothy A. Bates, Joon-Yong Lee, Thomas O. Metz, Carsten Schultz, and Fikadu G. Tafesse. 2022. “A Global Lipid Map Reveals Host Dependency Factors Conserved across SARS-CoV-2 Variants.” Nature Communications 13 (1): 3487. https://doi.org/10.1038/s41467-022-31097-7. https://www.nature.com/articles/s41467-022-31097-7.