The current study, which was based on random forest classification (RFC) modeling, revealed that the reduced abundance of Prevotella salivae and the metabolic pathways associated with lipopolysaccharide (LPS) and mycolic acid biosynthesis are the strongest predictors of COVID-19 patients who could require respiratory support in the form of supplemental oxygen or mechanical ventilation. These findings suggest that monitoring the composition of the oropharyngeal microbiome in the acute phase of COVID-19 may determine those who are at risk of suffering from severe disease.
Study: Oropharyngeal microbiome profiled at admission is predictive of the need for respiratory support among COVID-19 patients. Image Credit: crystal light / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
As of March 7, 2022, COVID-19, which is caused by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused over six million deaths worldwide. Clinical factors associated the with risk of developing severe disease include advanced age, obesity, and underlying comorbidities such as diabetes and high blood pressure. However, the individual factors that determine a patient’s response to COVID-19 may play a role in determining the requirement of hospitalization and respiratory support are not yet known.
Upper respiratory tract microflora has been reported to influence the host immune response to respiratory infections, such as certain commensal bacteria in the nasopharynx protect against influenza. Thus, the current study aims to investigate if the information on the oropharyngeal microbiome, combined with certain clinical factors, could predict the clinical trajectory of COVID-19 patients.
About the study
The team detected the oropharyngeal microbiome of COVID-19 patients using metagenomic DNA sequencing performed on oropharyngeal swabs. The study cohort consisted of 74 COVID-19-positive patients, of which 50 were acute cases with a known symptom duration of fewer than 14 days. Thirty-eight (76%) of these acute cases required some form of respiratory support.
Patient clinical characteristics at admission such as age and medical comorbidities were combined with the abundance of bacterial species and metabolic pathways. A machine learning model was then trained to determine which features, including bacterial species and metabolic pathway abundances, could predict the need for respiratory support.
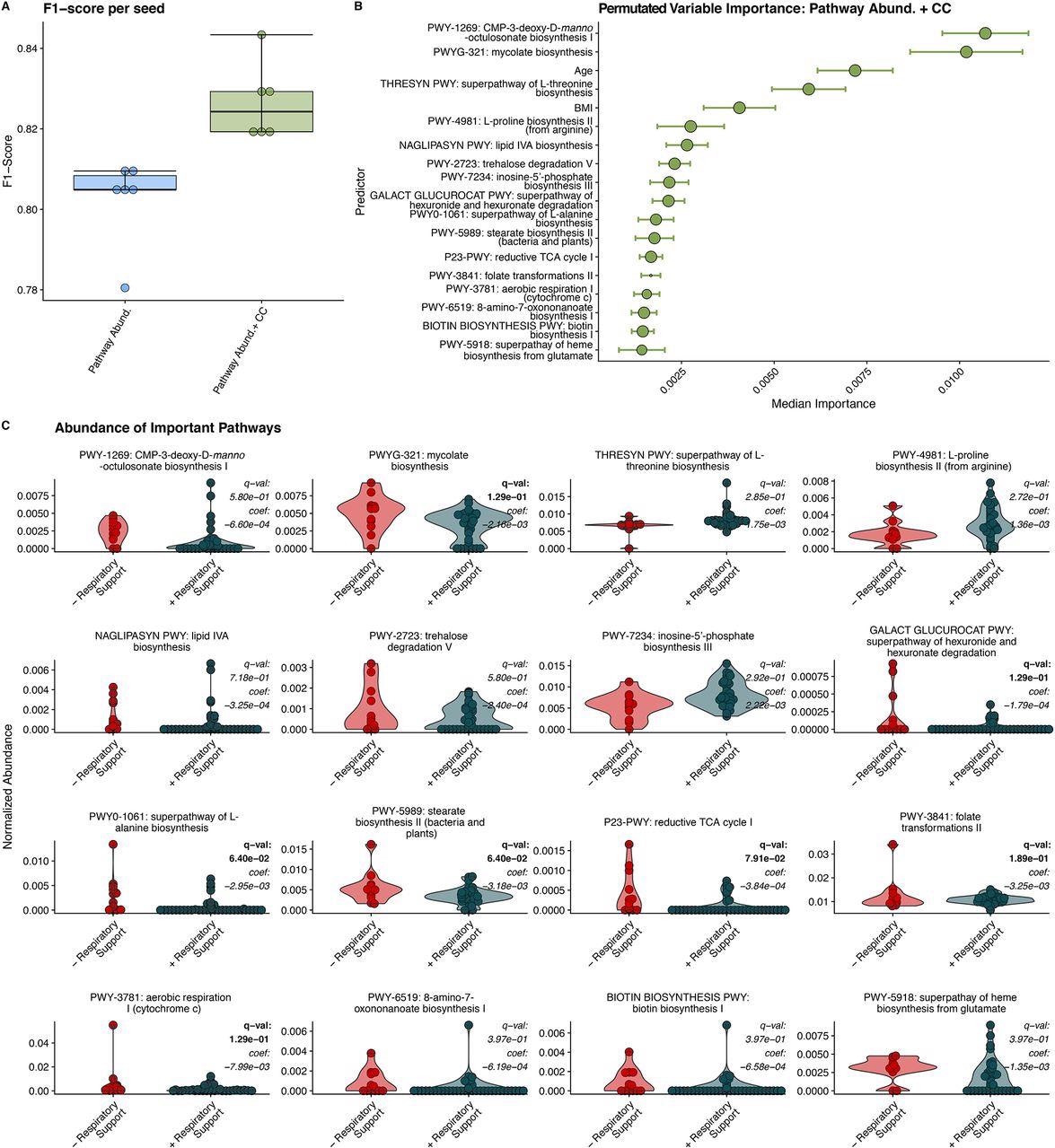
Random Forest Classification Using Metabolic Pathways. A) F1 scores of RFC models built on relative abundance of detected metabolic pathways and clinical covariates (CC). B) Median relative importance of variables in predicating the need for respiratory support within the trained with relative pathway abundances and clinical covariates (median importance ± median absolute deviation). C) Relative abundance of detected metabolic pathways in individuals requiring respiratory support and those not requiring respiratory support. MaAsLin2 derived q-values and coefficients are displayed for each pathway. Significant q values (q < 0.25) are bolded.
Study findings
The relative abundance of bacterium P. salivae, which is a gram-negative anaerobic organism and common oropharyngeal colonizer, was identified as the most important predictor of the need for respiratory support.
More specifically, a reduction in the abundance of P. salivae was indicative of respiratory support need. This suggested that the presence of these protective commensals was associated with COVID-19 patients not requiring respiratory support. Moreover, this organism was ranked to be even more predictive than the patients’ age and BMI, both of which are clinical factors that are associated with severe COVID-19.
Reduced abundance in other bacterial populations including Campylobacter concisus, Veillonella infantum, and Actinomyces species S6-Spd3 were also predictive of the need for respiratory support. All results from RFC modeling were validated by using independent analysis based on the general linear model MaAsLin2.
These findings suggest the beneficial effect of oropharyngeal bacteria against severe COVID-19 and corroborate with previous observational data regarding the treatment of COVID-19 patients with antibiotics. Previous data, combined with the current findings, suggest that the treatment of COVID-19 patients with antibiotics is potentially harmful due to their effect on beneficial commensal flora.
Together, these findings suggest that the presence of beneficial commensal bacteria in the upper airway has the potential to prevent or mitigate pulmonary manifestations of COVID-19."
Further, the team profiled the abundance of microbial metabolic pathways from sequencing data using the HMP Unified Metabolic Analysis Network (HUMAnN3). To this end, the top predictor metabolic pathways for the need for respiratory support included a reduced abundance of LPS biosynthesis (CMP-3-D-manno-octulosonate and lipid IV A biosynthesis) and mycolic acid biosynthesis pathways, followed by the trehalose degradation pathway. In addition, an increased abundance of L150 threonine, L-proline, as well as inosine-5-phosphate pathways and genes associated with aerobic utilization of hexuronides were also found to be predictive of respiratory support requirements.
Conclusions
A higher abundance of LPS biosynthetic pathways in the oropharynx appears to be protective against respiratory support requirements in COVID-19 patients. LPS is highly inflammatory; therefore, the likely translocation of inflammatory LPS-producing organisms from the oropharynx to the lungs might heighten inflammation during COVID-19 lung disease, thereby leading to the need for respiratory support.
This suggests that the oropharyngeal microbiome affects disease course in COVID-19 and could be targeted for diagnostic purposes to determine who may need oxygen, or therapeutic purposes such as probiotics to prevent severe COVID-19 disease manifestations.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bradley E. S., Zeamer, A. L., Bucci, V., et al. (2022) Oropharyngeal microbiome profiled at admission is predictive of the need for respiratory support among COVID-19 patients. medRxiv. doi:10.1101/2022.02.28.22271627. https://www.medrxiv.org/content/10.1101/2022.02.28.22271627v1.
- Peer reviewed and published scientific report.
Bradley, Evan S., Abigail L. Zeamer, Vanni Bucci, Lindsey Cincotta, Marie-Claire Salive, Protiva Dutta, Shafik Mutaawe, et al. 2022. “Oropharyngeal Microbiome Profiled at Admission Is Predictive of the Need for Respiratory Support among COVID-19 Patients.” Frontiers in Microbiology 13 (September). https://doi.org/10.3389/fmicb.2022.1009440. https://www.frontiersin.org/articles/10.3389/fmicb.2022.1009440.