Throughout the course of the coronavirus disease 2019 (COVID-19) pandemic, researchers around the world have worked towards understanding the underlying mechanisms associated with the transmission, ability to evade immunity, and many other aspects of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
T-cells play an important role in preventing severe SARS-CoV-2 infection. Although SARS-CoV-2 variants, particularly variants of concern (VOCs), contain multiple mutations that allow them to evade the immune response induced by either vaccination or natural infection, most T-cell epitopes remain conserved.
Thus, T-cell responses largely remain unaffected by mutations. Most importantly, SARS-CoV-2 memory T-cells persist for a prolonged period, thereby offering a greater degree of protection against both the wild-type SARS-CoV-2 strain its VOCs.
The frequency and magnitude of antigen-specific memory T-cells have a promising impact on COVID-19 manifestation. Therefore, it is imperative to understand the factors that contribute to the magnitude and breadth of the T-cell response during SARS-CoV-2 infection.
Previous studies have reported CD4+ and CD8+ T-cell activation to be inversely correlated with COVID-19 severity and death. These studies have further shown that a strong Th1-biased response is critical for SARS-CoV-2 recovery.
Additionally, the role of antibodies, particularly the immunoglobulin G (IgG) response against the SARS-CoV-2 receptor-binding domain (RBD), is related to patient survival. Scientists have also revealed that the recognition of viral nucleic acids by toll-like receptors (TLRs) on innate immune cells is critical for producing a suitable immune response to viral infection.
For the production of effective COVID-19 vaccines and therapies, it is important to understand the mechanisms associated with the induction of strong innate, humoral, and cellular responses against SARS-CoV-2.
About the study
In a new study published on the bioRxiv* preprint server, researchers investigate innate and adaptive immune responses at both the early and late stages of SARS-CoV-2 infection. The researchers were also interested in establishing the relationship between the two stages of infection and the immune responses during mild COVID-19 infection.
In their study, the researchers performed a longitudinal metabolic analysis of SARS-CoV-2-infected patients in agreement with the immune phenotype to reveal distinct metabolites that positively correlate with the innate and adaptive immune responses.
Previous studies have revealed that TLR signaling could be essential for inducing an innate immune reaction to SAR-CoV-2, which would consequently elicit a robust T-cell response. In the current study, scientists used TLR 3, 7, and 8 agonists to trigger peripheral blood mononuclear cells (PBMCs) from COVID-19 patients to determine their phenotypes in response. Herein, the researchers revealed that COVID-19 patients exhibit altered metabolic pathways and dysregulated energy production.
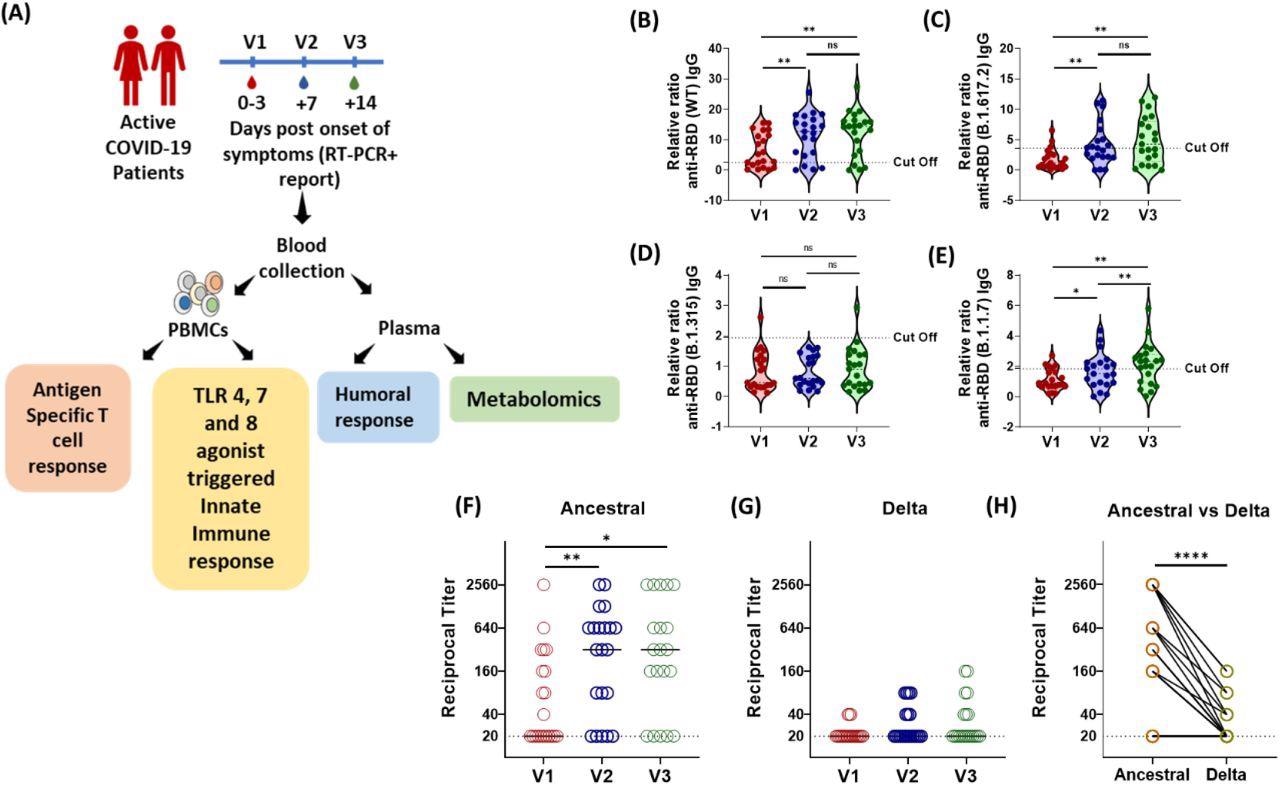
Longitudinal Analysis of Humoral Immune Response against the SARS-CoV-2 during acute COVID-19 infection. (A) Schematic experimental design and longitudinal sample collection at specific time intervals after confirmed SARS-CoV-2 infection in mildly infected patients. (B-E) The longitudinal anti-RBD IgG responses were evaluated by performing ELISA against the RBD proteins of (B) WT (Wuhan isolate) (C) Delta (B.1.617.2) (D) Beta (B.1.315) (E) Alpha (B.1.1.7). All data, represented as ratio-converted ELISA reads to a pool of pre-pandemic negative control samples (relative ratio), were plotted using violin plots. (F-H) The longitudinal neutralizing antibody titers against (F) the ancestral strain and (G) the delta strain of SARS-CoV-2 during V1 (day 0-3), V2 (day 7) and V3 (day 14) from COVID-19 positivity (H) The paired representation of NAb titers in the active COVID-19 patients during V3 against the ancestral and delta variants of SARS-CoV-2. The dots represent each individual sample. Tukey’s multiple comparisons and two-sided Wilcoxon Signed Rank tests were employed for unpaired and paired analysis, respectively. n.s = not significant, *= p 0.05, **= p 0.01, **** = p < 0.0001.
Study findings
The authors obtained samples during the first wave of COVID-19, before the emergence of the Delta and Omicron VOCs. This helped the researchers to determine how T-cells associated with the original SARS-CoV-2 strain interacted with the spike protein of the SARS-CoV-2 Delta and Omicron variants.
To analyze the humoral immune response, researchers tested the IgG response against the RBD proteins of the original SARS-CoV-2 strain and its VOCs. To this end, a sharp reduction in the cross-reactivity of RBD-specific IgG and neutralizing antibodies was observed against COVID-19 patients infected by VOCs.
The frequency of activated induced markers in CD4+ and CD8+ T-cells were then assessed and found to be consistent with previous reports. A Th1-skewed response has been deemed to be a signature of anti-SARS-CoV-2 T-cell immunity.
In this study, early-stage T-cells were found to primarily express interferon γ (IFN-γ), rather than interleukin 2 (IL-2). Moreover, IL-2 was not found to be primarily involved in the early T-cell response; however, this cytokine might be dominant in the later stages of SARS-CoV-2 infection.
The T-cell response elicited during the active infection persists; however, its function is reduced against the Delta and Omicron infection as compared to the original SARS-CoV-2 strain. Moreover, the overall magnitude of the T-cell response was low in acute samples, which indicates a limited scope for cross-reactivity against the VOCs. Additionally, the authors revealed that the antiviral TLR specific proinflammatory cytokine activity by PBMCs could play an important role in generating a strong immune response against COVID-19.
The metabolic state that modulates the immune response, and vice versa, was assessed by conducting a plasma metabolomic analysis of COVID-19 patients. To this end, a significant correlation between the arginine biosynthesis metabolites and SARS-CoV-2 specific immune responses was observed.
Additionally, an increase in norvaline, which is an arginase inhibitor, during COVID-19 was observed, which is related to the CD8+ T-cell response. The researchers reported reduced citrate and aconitate levels in COVID-19 patients.
Conclusions
One of the limitations of this study is its small sample size. Additionally, the researchers failed to determine type I IFN levels upon TLR stimulation of PBMCs.
The longevity of T-cell immune responses was also not determined. However, the current study indicated that PBMC-derived innate immune responses are essential for producing a robust T-cell response.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Binayke, A., Zaheer, A., Dandotiya, J., et al. (2022) Innate immune response and distinct metabolomic signatures together drive and shape the SARS-CoV-2-specific T cell response during COVID-19. bioRxiv. doi:10.1101/2022.03.11.483930. https://www.biorxiv.org/content/10.1101/2022.03.11.483930v1.
- Peer reviewed and published scientific report.
Binayke, Akshay, Aymaan Zaheer, Jyotsna Dandotiya, Sonu Kumar Gupta, Shailendra Mani, Manas Ranjan Tripathy, Upasna Madan, et al. 2022. “Proinflammatory Innate Cytokines and Distinct Metabolomic Signatures Shape the T Cell Response in Active COVID-19.” Vaccines 10 (10): 1762. https://doi.org/10.3390/vaccines10101762. https://www.mdpi.com/2076-393X/10/10/1762.