Study: Whole-genome risk prediction of common diseases in human preimplantation embryos. Image Credit: Explode / Shutterstock.com
Preimplantation genetic testing
Preimplantation genetic testing of in vitro fertilized embryos allows for researchers to identify the presence of any potential genetic disorders before implantation. Genetic testing is currently used to prevent rare Mendelian disorders.
Some studies have also tested embryos for heart disease and cancers. These studies have used a polygenic risk score (PRS), wherein the effects of multiple genetic variants are combined into a single predictor.
The DNA material for genetic testing is acquired from single-cell or few-cell embryo biopsies, which can limit the quantity and quality of DNA for sequencing. Thus, comprehensive genome profiling of embryos is cost- and time-intensive. Moreover, inaccuracies in genome profiles can arise due to several technical reasons, which can also prevent the accurate detection of rare gene mutations.
Whole-genome reconstruction
In the current study, 110 embryos were obtained from ten couples who underwent in vitro fertilization (IVF) with prior clinical preimplantation genetic testing. DNA was available from ten children born through IVF, all of whom also had preimplantation genetic testing results. Preimplantation genetic testing for aneuploidy results showed that 68 embryos were euploid and 42 embryos had one or more aneuploid chromosomes.
Whole-genome reconstruction of embryos was achieved by genome sequencing both parents and array measurements of the sibling embryos. A combination of molecular and statistical approaches was used to link the parental variants into ‘haplotypes’ that corresponded to the individual chromosomes.
The locations of meiotic recombination for each embryo were determined. Additionally, relevant haplotype segments were assembled to approximate the entire inherited embryonic genome.
The accuracy of the whole-genome reconstruction was evaluated by
comparing the predicted genotypes to the genotypes of the born children. An average of 5.8 million sites in embryos, ranging from 5.4 to 6.4 million variants, were predicted. The prediction accuracy was 96.3–98.4% in day three embryo biopsies and 98.0–98.9% in day five embryo biopsies as compared to the born child.
In four couples, Transposase Enzyme-Linked Long-read Sequencing (TELL-Seq) was performed for library preparation. This captured the rare variants increasing the number and accuracy of rare-variant predictions in each family.
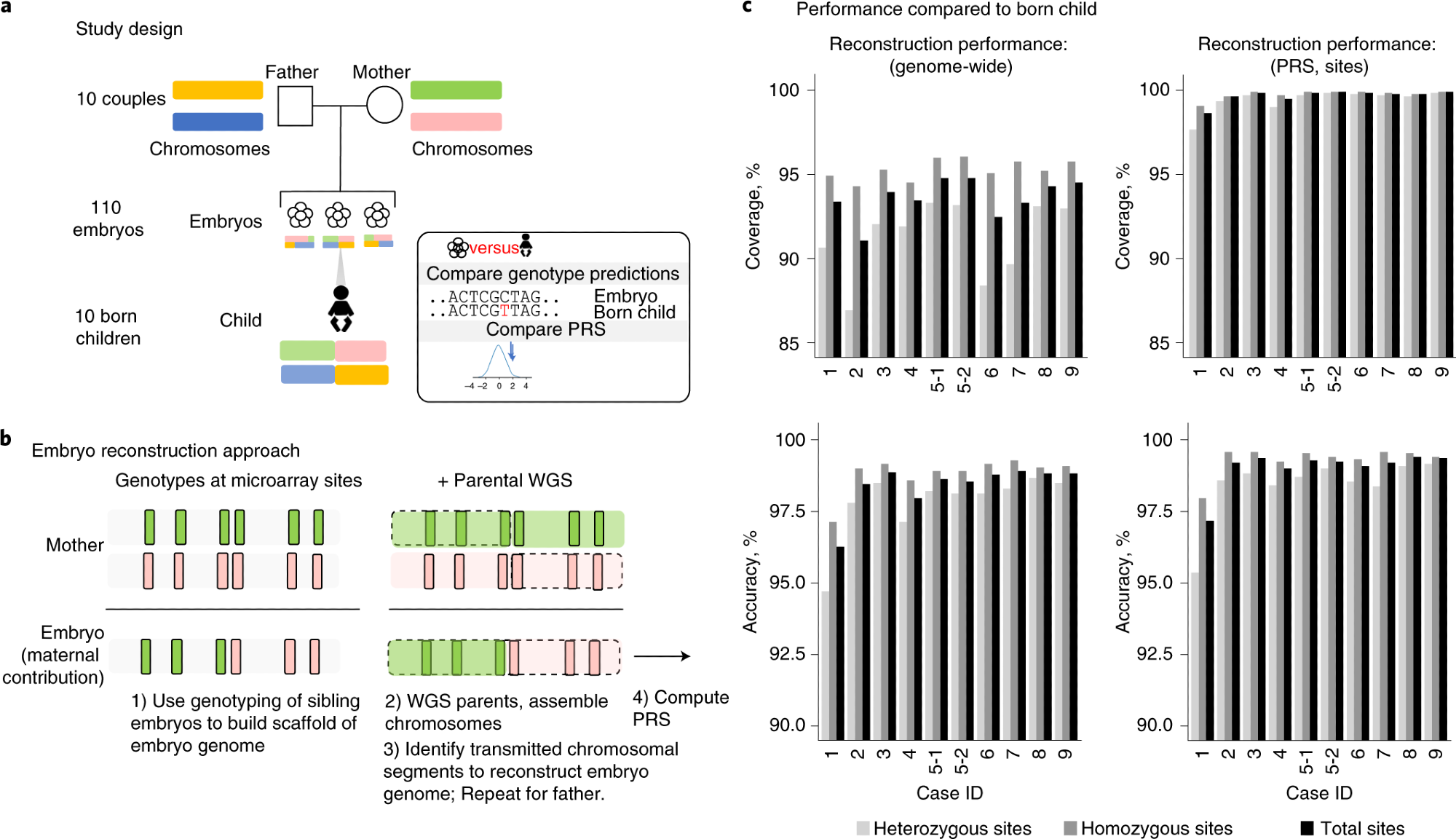 a, This research study involved reconstruction of 110 embryo genomes from 10 couples and comparison to the genome sequence of the born child. Twelve PRS models were computed from the born-child samples and the 10 corresponding reconstructed embryos and compared for concordance. b, WGR involves whole-genome sequencing (WGS) of prospective parents and single-nucleotide polymorphism (SNP) microarray genotyping of sibling embryos. Allele measurements at each SNP are color-coded based on the parental haplotype of origin illustrated in a. A combination of molecular and statistical/population-based techniques phase the parents’ chromosomes infer the locations of meiotic recombination for each embryo and correct errors introduced in the process of testing single-cell or few-cell embryo biopsies. Reconstructed embryo whole genomes are used to predict common disease risk by calculating PRSs and inferring the inheritance of rare variants with high impact on disease risk. c, Performance by comparing genotypes from WGR with the born child’s DNA shows genotype accuracies ranging from 99.0% to 99.4% at sites used in polygenic prediction in day-5 embryos and 97.2% to 99.1% in day-3 embryos. Case 1 includes only day-3 embryos, and case 2 includes both day-3 and day-5 embryos. All other cases included day-5 embryos only. Statistics are subdivided by genotype (heterozygous or homozygous) in the born child.
a, This research study involved reconstruction of 110 embryo genomes from 10 couples and comparison to the genome sequence of the born child. Twelve PRS models were computed from the born-child samples and the 10 corresponding reconstructed embryos and compared for concordance. b, WGR involves whole-genome sequencing (WGS) of prospective parents and single-nucleotide polymorphism (SNP) microarray genotyping of sibling embryos. Allele measurements at each SNP are color-coded based on the parental haplotype of origin illustrated in a. A combination of molecular and statistical/population-based techniques phase the parents’ chromosomes infer the locations of meiotic recombination for each embryo and correct errors introduced in the process of testing single-cell or few-cell embryo biopsies. Reconstructed embryo whole genomes are used to predict common disease risk by calculating PRSs and inferring the inheritance of rare variants with high impact on disease risk. c, Performance by comparing genotypes from WGR with the born child’s DNA shows genotype accuracies ranging from 99.0% to 99.4% at sites used in polygenic prediction in day-5 embryos and 97.2% to 99.1% in day-3 embryos. Case 1 includes only day-3 embryos, and case 2 includes both day-3 and day-5 embryos. All other cases included day-5 embryos only. Statistics are subdivided by genotype (heterozygous or homozygous) in the born child.
Polygenic risk score
Whole-genome reconstruction allowed for the prediction of variants in the embryo genomes. For predicting the common disease risk by combining the variants, polygenic risk scores were calculated using the embryo genomes.
For each embryo, risk scores were calculated for a set of published polygenic models and were subsequently validated and calibrated in the U.K. Biobank (UKB) on a population-specific basis. The genotype accuracy at sites relevant to polygenic risk scoring was 97.2–99.1% in cases from day three embryo biopsies and 99.0–99.4% in cases from day five embryo biopsies as compared to high-confidence positions from the whole-genome sequences of the born children.
The polygenic risk scores of each embryo were normalized and converted into predicted odds of disease using a statistical logistic regression model. There was a high correlation between the computed polygenic risk scores from embryo biopsies and those computed from samples of born children. Variability in polygenic disease risk in embryos was observed from different families and in sibling embryos within families.
Rare variant identification
Combining rare variants with polygenic risk scores magnified predicted differences across sibling embryos. In a couple with a pathogenic BRCA1 variant, the predicted genetic risk for breast cancer varied 15-fold across the embryos.
Using rare variant BRCA1 or polygenic risk score alone, the predicted genetic risk for breast cancer varied 4.5-fold or three-fold across the embryos. Preimplantation genetic testing for breast cancer is most beneficial when the parents carry a pathogenic variant.
One couple was incidentally found to carry an APC risk allele, an established colon cancer gene. One out of three embryos of this couple was predicted to harbor the APC risk allele.
Importantly, the BRCA1 and APC rare variants identified in this study would have been missed by the current methods.
Future directions
The approach used in the current study is limited to inherited genetic variations and, therefore, excludes new variations that may arise after conception. Though such new variations are rare, they represent a significant portion of early-onset neurodevelopmental disorders like autism and intellectual disability.
Therefore, it is important to detect these variations, which will be possible along with ultradeep sequencing of embryo biopsies. Thus, the approach described here provides better predictions in day five embryos rather than with day three embryos.
The risk scores calculated in this study are valid in European populations because human genetics research data is available for individuals of European ancestries. For non-Europen populations, the predictions may not be accurate.
Additional research is needed to confirm the applicability of this approach in clinical settings, as these findings may not be generalizable to in vitro fertilized sibling embryos. Thus, there is a risk that prediction accuracy may be lower in clinical settings.
Studies on human embryos raise several ethical considerations. Using polygenic information for prenatal decision-making will need further ethical and practical considerations. In a survey in the U.S., 68% of 1,457 participants believed the reasonability of using embryo screening for PRS.
A proportion of participants in this study were referred to perform PGT-A for aneuploidy screening. PGT-A is under debate for its clinical effectiveness. The current study does not address the accuracy of predictions in mosaic or aneuploid embryos.
Conclusions
The current study opens an avenue for discussing the clinical utility and practice of genome-based preimplantation genetic testing.
With the advent of cost-effective sequencing technology and a better understanding of risk scores, this approach may provide reliable risk predictions to couples undergoing IVF, especially those with a personal or family history of common diseases.
Journal reference:
- Kumar, A., Im, K., Banjevic, M., et al. (2022). Whole-genome risk prediction of common diseases in human preimplantation embryos. Nature Medicine 28(3); 513–516. doi:10.1038/s41591-022-01735-0.