The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) readily transmits in close contact through respiratory droplets. The nasal cavity is the primary site of viral entry; thus, SARS-CoV-2 infection of the upper airway is an essential factor in the pathogenesis of coronavirus disease 2019 (COVID-19).
 Study: Evolution of nasal and olfactory infection characteristics of SARS-CoV-2 variants. Image Credit: Lightspring / Shutterstock.com
Study: Evolution of nasal and olfactory infection characteristics of SARS-CoV-2 variants. Image Credit: Lightspring / Shutterstock.com

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
In the present study, researchers evaluated changes in the olfactory and nasal epithelial epithelium in infections caused by the SARS-CoV-2 variants.
To assess the cellular tropism in the nasal mucosa, WA1 strain infections in vitro were performed using human nasal tissues, which were discarded post skull base and endonasal sinus surgeries. The antibodies were screened to visualize the nucleocapsid (NP) or spike (S) proteins in seven COVID-19 tissue explants.
Antibodies were also immunostained and SARS-CoV-2 ribonucleic acid (RNA) was detected using an RNAscope. The neuronal marker Tuj1 was used for verifying the changes in the olfactory epithelium, which was stained with viral NP, and NP+ cells were counted.
Cellular tropism of the SARS-CoV-2 WA1, Omicron, and Delta strains in the olfactory and respiratory nasal mucosa was evaluated using hamsters and validated by measuring the angiotensin-converting enzyme 2 (ACE2) messenger ribonucleic acid (mRNA) expression using quantitative-polymerase chain reaction (qPCR) analysis in C57BL/6J mice at two weeks, two months, and 19 months. Post viral inoculation with a median tissue culture infectious dose (TCID50) of 1×105, confocal images were captured.
After establishing the high tropism of Delta and WA1 in olfactory mucosae, WA1 and Delta infections in olfactory neurons were assessed. For this, SARS-CoV-2 was intranasally inoculated into young and adult hamsters and uniform SARS-CoV-2 infections were generated in C57BL/6J mice using the mouse-adapted SARS-CoV-2 (maSARS) model to evaluate age-wise variations.
Study findings
A 700-times greater ACE2 expression was detected in sustentacular cells of the olfactory epithelium as compared to the respiratory epithelium. The immunostained antibodies predominantly present in the apical sustentacular cells showed staining patterns consistent with viral RNA.
In the WA1-positive human samples of young persons under the age of 30 years, the viral load in Tuj1+ neurons and axon bundles was remarkably increased. WA1 infection of the olfactory neurons and olfactory bulb transport through axons was also profound in young individuals.
Delta infection demonstrated a greater submucosal invasion capacity, while the SARS-CoV-2 Omicron variant demonstrated greater retention in the sinonasal epithelial cells. In contrast to the WA1 or Delta strain, Omicron-infected olfactory epithelium significantly reduced to 6.7%, which was indicative of lower Omicron pathogenicity.
Pronounced NP+ sinonasal cells at a concentration of 3.3 positive cells/mm of epithelium were detected in Omicron hamsters, thus indicating a longer duration of Omicron infections. Omicron-infected NP+ cells in the sinus and nasal mucosa increased by seven- to ten-fold as compared to WA1 or Delta, thus indicating a transitional shift in tropism from the olfactory epithelium to respiratory epithelium with viral evolution.
In maSARS animals, a substantial cluster of differentiation 45 (CD45+) immune infiltration was noted in the lamina propria and neuroepithelium, thereby indicating nasal immune defense to SARS-CoV-2 infection.
At four days post-infection (dpi) in hamsters, viral inoculation induced extensive shedding of NP+ cells into the nasal lumen, thus indicating viral clearance by macrophages. In addition, widespread distribution of Iba1+ macrophages was detected in the olfactory mucosa, of which 72% were also NP+.
At six dpi, 48% of CD45+ cells in the olfactory mucosa were Iba1+ macrophages, 3.7-times higher in the nasal lumen in older individuals as compared to young ones, which was indicative of delayed viral clearance in older COVID-19 patients. In the single-cell RNA sequencing (scRNA-seq) dataset analysis, a significant reduction in phagocytosis-related genes such as Clec4n (Dectin2), Fpr2, Cd9, and Fabp5 was observed in old macrophages as compared to young mice. This indicated that macrophages are essential for immune defenses and their decreased capacity for viral clearance could prolong SARS-CoV-2 retention in olfactory mucosae of the older COVID-19 patients.
A 100- to 300-fold increase in NP+ cells was observed in the Tuj1+ olfactory epithelium as compared to the Tuj1-negative respiratory epithelia. Most NP+ cells showed co-localization with Krt18+ sustentacular (olfactory) cells.
Corresponding structural deterioration was observed in olfactory mucosa (high viral load) but not in respiratory epithelium (low viral load). These findings were indicative of the olfactory-specific WA1 tropism, which is responsible for anosmia in COVID-19 patients.
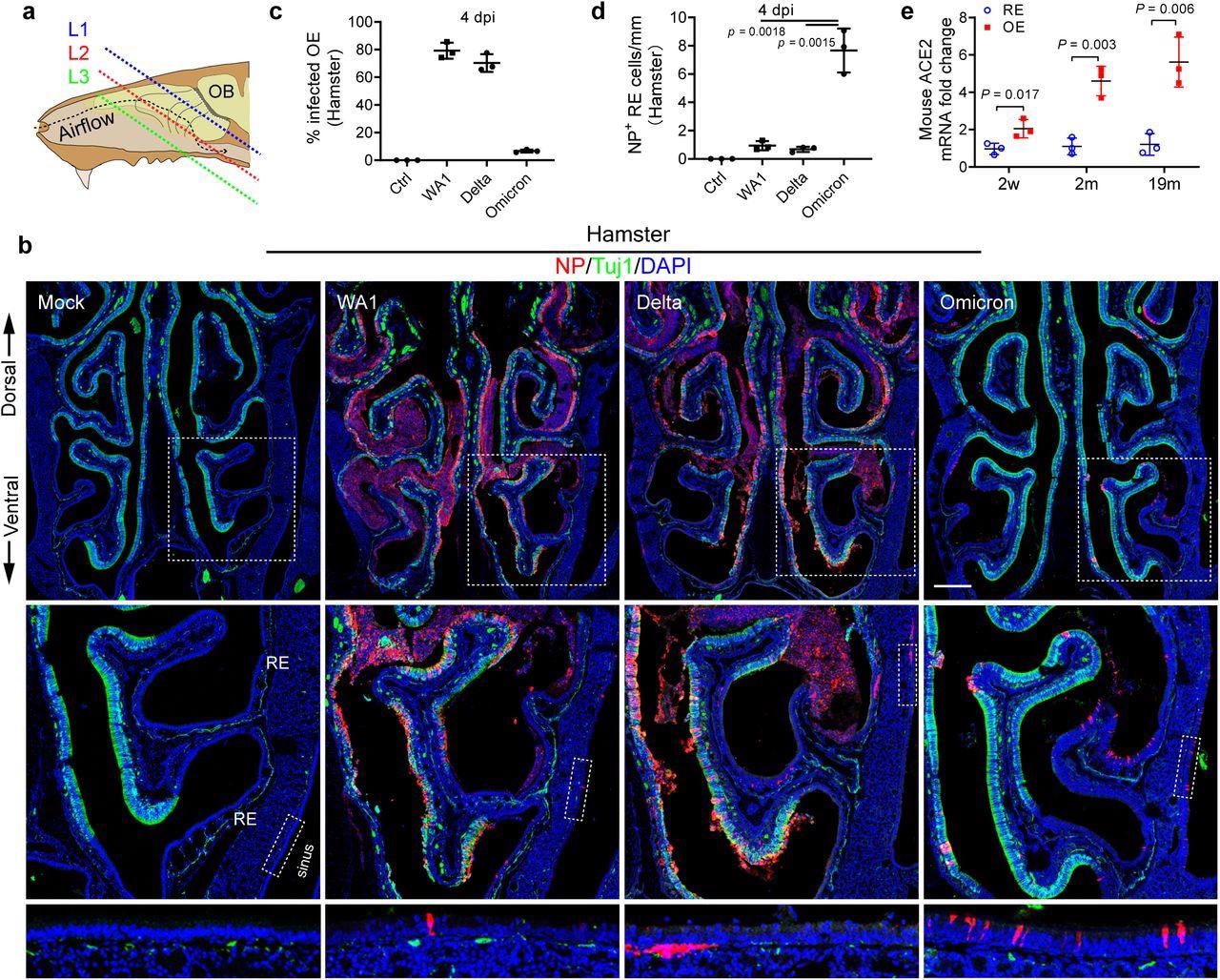 Omicron variant shows tropism transition from olfactory to respiratory epithelium. a, Scheme of the tissue section. To avoid variability across different animals, frozen sections were collected and examined at three consistent levels (L1–3) representing the anterior (mainly respiratory epithelium), middle (respiratory + Olfactory epithelium), and posterior (mainly olfactory epithelium). b, Confocal images of NP and Tuj1-labeled hamster nasal sections at L2. WA1, Delta, and Omicron infected hamsters were examined on 4 dpi. Boxed areas are highlighted at bottom. Scale bars = 500 µm. c, Percentage of the infected olfactory epithelium. The total length of Tuj1+ or NP+/Tuj1+ epithelium in each section at L1-3 were quantified using Image J. d, Quantification of NP+ cells in nasal respiratory epithelium. The total NP+ cells in Tuj1- respiratory epithelium including paranasal sinuses of each section were counted. e, qPCR analysis of ACE2 expression in mouse nasal respiratory or olfactory epithelium at age of 2 weeks, 2months, and 19months. The entire nasal respiratory or olfactory epithelium from the same animal were isolated separately. Data are represented as mean ± S.D. Statistical significance was determined by unpaired two-tailed t-test. Each data point represents an individual animal.
Omicron variant shows tropism transition from olfactory to respiratory epithelium. a, Scheme of the tissue section. To avoid variability across different animals, frozen sections were collected and examined at three consistent levels (L1–3) representing the anterior (mainly respiratory epithelium), middle (respiratory + Olfactory epithelium), and posterior (mainly olfactory epithelium). b, Confocal images of NP and Tuj1-labeled hamster nasal sections at L2. WA1, Delta, and Omicron infected hamsters were examined on 4 dpi. Boxed areas are highlighted at bottom. Scale bars = 500 µm. c, Percentage of the infected olfactory epithelium. The total length of Tuj1+ or NP+/Tuj1+ epithelium in each section at L1-3 were quantified using Image J. d, Quantification of NP+ cells in nasal respiratory epithelium. The total NP+ cells in Tuj1- respiratory epithelium including paranasal sinuses of each section were counted. e, qPCR analysis of ACE2 expression in mouse nasal respiratory or olfactory epithelium at age of 2 weeks, 2months, and 19months. The entire nasal respiratory or olfactory epithelium from the same animal were isolated separately. Data are represented as mean ± S.D. Statistical significance was determined by unpaired two-tailed t-test. Each data point represents an individual animal.
Olfactory epithelium could be identified based on Tuj1+ staining, increased neuroepithelial thickness, and the presence of axon bundles. In nine-month-old animals, ACE2 mRNA levels were higher in the adult olfactory epithelium by five- to seven-fold when compared to the respiratory epithelium, thus indicating that the ACE2 expression pattern underpins the olfactory epithelium as a viral replication site.
Low ACE2 expression in olfactory neurons indicated the mediation of neuronal entry by other receptors such as neuropilin-1 (Nrp1). Over 30% and 10% of Tuj1+ olfactory neurons expressed Nrp1 in young and adult mice, respectively, which is indicative of an age-wise reduction in olfactory neuronal infection and axonal transport.
NP+ axons were present in the olfactory nerve layer (ONL) in young hamsters, thus indicating viral transport to the olfactory bulb. Additionally, Iba1+ microglial cells in young olfactory bulbs were 1.7-times higher when compared to older hamsters.
Conclusions
The current study demonstrated an age-associated increase in the rate of SARS-CoV-2 infection in the olfactory epithelium as compared to the respiratory epithelium of human nasal mucosa. A transition from the olfactory to the respiratory epithelium was observed with viral evolution, which explains the low prevalence of anosmia in Omicron infections.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.