The SARS-CoV-2 pandemic continues via the consecutively emerging viral variants. Various coronavirus disease 2019 (COVID-19) vaccines have been established centered on the initial SARS-CoV-2 Wuhan strains. Fortuitously, they have been effective against the subsequently aroused viral strains.
The number of SARS-CoV-2 infections dropped in certain nations, perhaps as a result of the effectiveness of the vaccination. Nonetheless, the global COVID-19 pandemic has not yet been contained.
Antiviral therapy is efficacious during the SARS-CoV-2 replication stage, occurring in the early phases of infection. Similarly, using therapeutic neutralizing antibodies against COVID-19 has been significantly effective. Unfortunately, there exist few beneficial antibodies to combat evolving SARS-CoV-2 strains.
About the study
In the present research, the scientists developed multiple monoclonal antibodies from SARS-CoV-2-convalescent patients. Since the start of the SARS-CoV-2 epidemic in March 2020 in Japan, the authors have collected peripheral blood samples from recovering COVID-19 patients, which have been used to generate neutralizing antibodies.
The investigators procured blood samples from discharged SARS-CoV-2 patients from Keio University Hospital. The cell-based SARS-CoV-2 spike (S)-angiotensin-converting enzyme 2 (ACE2) inhibition test was used to evaluate the neutralizing capacity of sera. The team selected 12 patients exhibiting prominent neutralizing concentrations for antibody generation.
The authors profiled patient-stemmed antibodies using 1) S-ACE2 inhibition assessment and 2) the association between the attaching capacity of these antibodies towards S-expressing cells and their potential to hinder the attaching of ACE2 against S-expressing cells. To more thoroughly analyze these antibodies, they also investigated the neutralizing potential using a cell fusion experiment. The researchers conducted an end-point microneutralization screening to verify that the chosen antibodies could neutralize the authentic SARS-CoV-2.
To further identify potential antibodies, the scientists evaluated the affinity toward SARS-CoV-2 receptor-binding domain (RBD) antigen and analyzed the overlap of epitopes. They chose five antibodies and used a pseudovirus harboring the S protein of the original SARS-CoV-2 Wuhan sequence and four significant variants to conduct a neutralization experiment before variants of concern (VOCs) arose. After the emergence of VOCs, they tested the antibodies' capacity to neutralize the original WK-521 virus and its variants, including Beta, Alpha, Gamma, Kappa, Delta, and Omicron BA.2 and BA.1.
The team conducted a cryo-electron microscopy (cryo-EM) investigation to get a structural understanding of antibodies and the SARS-CoV-2 S protein. The currently discovered antibodies employed in the in vivo investigation possessed the N297A mutation in the immunoglobulin G1 (IgG1)-fragment crystallizable (Fc) region to avert antibody-dependent enhancement (ADE). In addition, N297A mutation decreases adherence to the Fc receptor. The investigators then examined the effects of these antibodies in two animal models (a cynomolgus macaque model and a hamster model) to see the impact of these antibodies in in vivo settings.
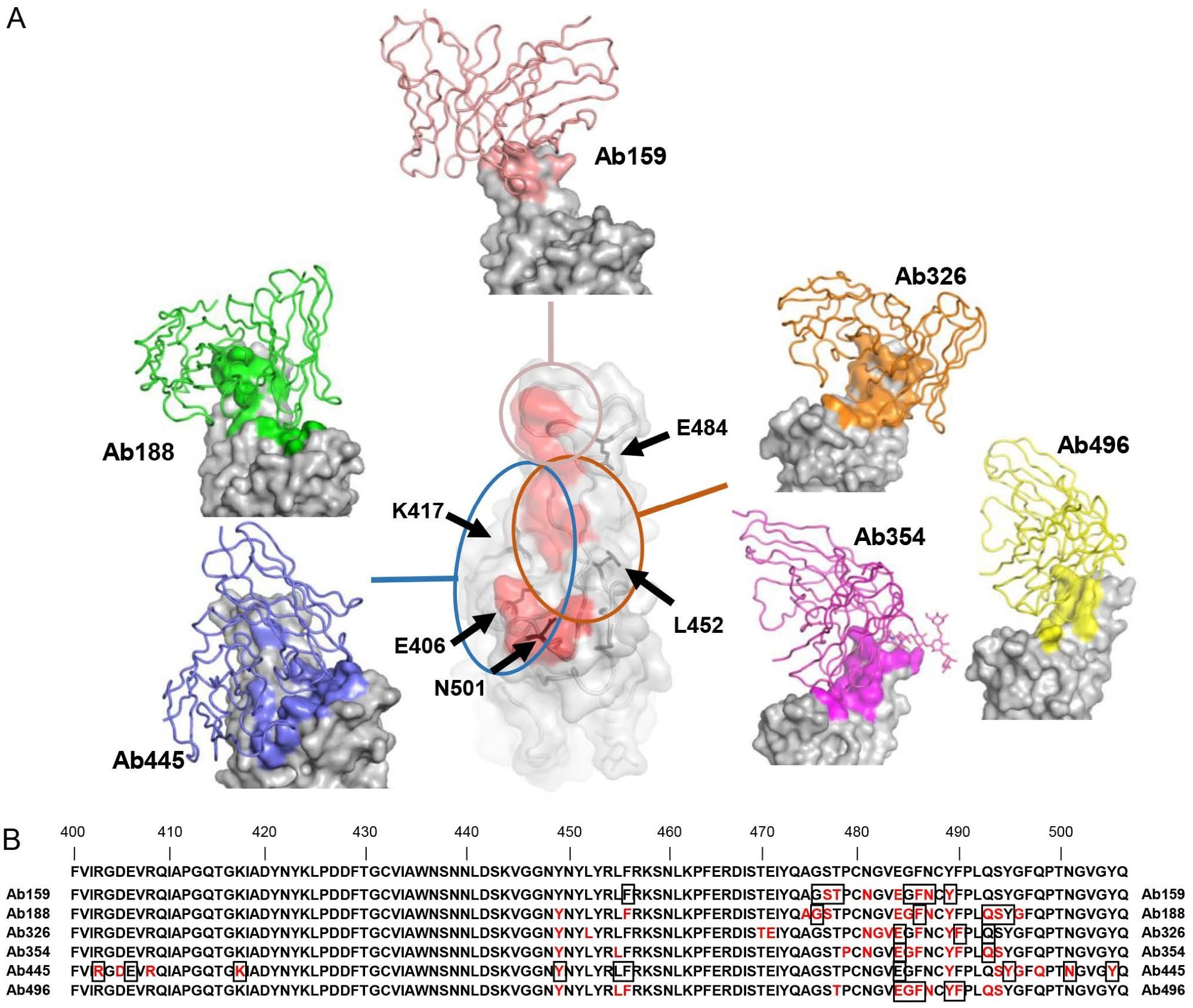 Cryo-EM structure of neutralizing antibodies (A) The structures of RBD and Ab159, Ab188, Ab326, Ab354, Ab445, and Ab496 are shown. Only the variable domains of antibodies are modeled and drawn as a cartoon tube (individual color) on the RBD surface (gray), and the epitope of each antibody is colored the same as each antibody. The red area in the central RBD is the binding residue of ACE2 (7A94) (Benton et al., 2020), showing the relationship between the binding sites of the antibodies, which are roughly divided into three groups. The positions of key amino acids are indicated by black arrows. (B) The residues 400-506 of Spike are shown.
Cryo-EM structure of neutralizing antibodies (A) The structures of RBD and Ab159, Ab188, Ab326, Ab354, Ab445, and Ab496 are shown. Only the variable domains of antibodies are modeled and drawn as a cartoon tube (individual color) on the RBD surface (gray), and the epitope of each antibody is colored the same as each antibody. The red area in the central RBD is the binding residue of ACE2 (7A94) (Benton et al., 2020), showing the relationship between the binding sites of the antibodies, which are roughly divided into three groups. The positions of key amino acids are indicated by black arrows. (B) The residues 400-506 of Spike are shown.
Results
The researchers discovered 494 antibodies from COVID-19-recovered persons, most showing an identical SARS-CoV-2 neutralizing ability to clinically employed antibodies in the neutralization assessment. Initially, antigen-specific memory B cells and antigen-nonspecific plasma cells were used to create antibodies. Nevertheless, the former harbored superior antibodies, highlighting the significance of choosing B cells by antigen. The data from the end-point authentic viral neutralization assay confirmed the findings from the cell-based S-ACE2 inhibition and cell fusion assays screening neutralizing antibodies.
Cryo-EM and cell-based mutant S-ACE2 inhibition experiments identified the epitopes on the S protein since antibodies were selected by competing with ACE2, classifying antibody attachment to S as class 1/2. N297 insertion on IgG1-Fc was one of the features of the antibodies discovered in this study. This mutation nearly eradicated the adhesion to Fc receptors. Indeed, it stopped Fc-facilitated uptake of the virus to Raji cells.
The selected antibodies were comparable to or better than imdevimab, a COVID-19 therapeutic agent, in neutralization testing against the Wuhan strain and VOCs utilizing authentic viruses and pseudoviruses. Regarding the in vivo activity of these antibodies, they showed potency for therapeutic usage in macaque and hamster models. Under doses of around 5 to 7 mg/kg, the current antibodies showed therapeutic efficacy in hamsters and macaques without causing a hike in viral uptake via ADE.
Conclusions
Overall, in the current study, the authors generated many antibodies from the B cells of convalescent COVID-19 patients infected by the SARS-CoV-2 D614G mutant or Wuhan strain. In addition, they identified numerous neutralizing antibodies with potent anti-SARS-CoV-2 variant strain neutralization properties.
These Fc-modified neutralizing antibodies from SARS-CoV-2 recovered persons had neutralizing properties comparable to clinical COVID-19 antibodies. The effectiveness of these antibodies was illustrated through infection research with macaque and hamster models in vivo and authentic viral and pseudovirus neutralization assays in vitro. These findings demonstrated that the currently discovered antibodies had adequate antiviral activity to serve as COVID-19 treatment options.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fc-modified SARS-CoV-2 neutralizing antibodies with therapeutic effects in two animal models; Masaru Takeshita, Hidehiro Fukuyama, Katsuhiko Kamada, Takehisa Matsumoto, Chieko Makino-Okamura, Tomomi Uchikubo-Kamo, Yuri Tomabechi, Kazuharu Hanada, Saya Moriyama, Yoshimasa Takahashi, Hirohito Ishigaki, Misako Nakayama, Cong Thanh Nguyen, Yoshinori Kitagawa, Yasushi Itoh, Masaki Imai, Tadashi Maemura, Yuri Furusawa, Hiroshi Ueki, Kiyoko Iwatsuki-Horimoto, Mutsumi Ito, Seiya Yamayoshi, Yoshihiro Kawaoka, Mikako Shirouzu, Makoto Ishii, Hideyuki Saya, Yasushi Kondo, Yuko Kaneko, Katsuya Suzuki, Koichi Fukunaga, Tsutomu Takeuchi. bioRxiv preprint 2022. DOI: https://doi.org/10.1101/2022.06.21.496751, https://www.biorxiv.org/content/10.1101/2022.06.21.496751v1
- Peer reviewed and published scientific report.
Takeshita, Masaru, Hidehiro Fukuyama, Katsuhiko Kamada, Takehisa Matsumoto, Chieko Makino-Okamura, Tomomi Uchikubo-Kamo, Yuri Tomabechi, et al. 2022. “Potent SARS-CoV-2 Neutralizing Antibodies with Therapeutic Effects in Two Animal Models.” IScience 25 (12): 105596. https://doi.org/10.1016/j.isci.2022.105596. https://www.cell.com/iscience/fulltext/S2589-0042(22)01868-5.