This female aged between 60 to 69 began receiving her cancer treatment at Shamir Medical Center hospital, Be'er Yaacov, Israel, in June 2021. Her oncological findings included malignant melanoma, diffuse large B-cell lymphoma, and squamous cell carcinoma. She also received treatment for fibromyalgia, asthma, and chronic obstructive pulmonary disease.
The patient contracted coronavirus disease 2019 (COVID-19) in September 2021 and remained persistently infected for over eight months despite receiving monoclonal antibody (mAb) cocktail treatments twice.
Typically healthy human hosts transmit SARS-CoV-2 for less than 14 days and do not transmit emerging mutations due to strong purifying selection. Conversely, the risk of persistent SARS-CoV-2 infection in immunocompromised individuals receiving the anti-cluster of differentiation 20 (CD20) medication rituximab is exceptionally high. Antiviral therapies, such as monoclonal/ polyclonal antibodies, ivermectin, and remdesivir, also do not always successfully treat such patients due to resistant viral mutants that emerge in some cases.
Subsequently, SARS-CoV-2 evolves more rapidly in such patients giving rise to mutations that increase viral fitness and enhance escape from neutralizing antibodies (nAbs). This could partially be the reason for the emergence of SARS-CoV-2 novel variants of concern (VOC), which are a threat to public health globally.
About the study
In the present study, researchers compared the mutational profile of the viral samples obtained from SARS-CoV-2 infected immunocompromised cancer patients with the samples representing the closest phylogenetic relative to identify mutations that emerged within her patient over the course of infection. They also compared emerging SARS-CoV-2 mutations in the presence and absence of nAbs.
The researchers tracked SARS-CoV-2 evolution in this patient for around six months, during which they obtained three nasopharyngeal samples (S1, S2, S3) for SARS-CoV-2 whole-genome sequencing (WGS). They estimated the global prevalence of various mutations using statistics from the global initiative on sharing all influenza data (GISAID) database. Further, the team captured the SARS-CoV-2 mutations within the patient and distinguished minor mutations with allele frequency (AF)<0.5 from major mutations, that became fixed and had AF>0.5.
They quantitatively determined anti-spike 1 and anti-spike 2 specific immunoglobulin G (IgG) antibodies in patient samples using a chemiluminescent immunoassay (CLIA) which uses magnetic beads coated with spike 1 and spike 2 antigens. Likewise, they used measurements of deep mutational scanning experiments to assess antibody escape.
Study findings
The WGS results revealed the patient was infected with the SARS-CoV-2 Delta variant, lineage AY.43. These viral samples were genetically correlated with contemporary viral genomes collected in Israel and Europe. The earliest sample S1 was more mutated than its nearest phylogenetic relative, with 10 new substitutions. The genomes from samples S2 and S3 showed a pattern of accumulating mutation, although their distribution was uneven. Seven emerging mutations accumulated in less than a month, while another 10 in the next three months.
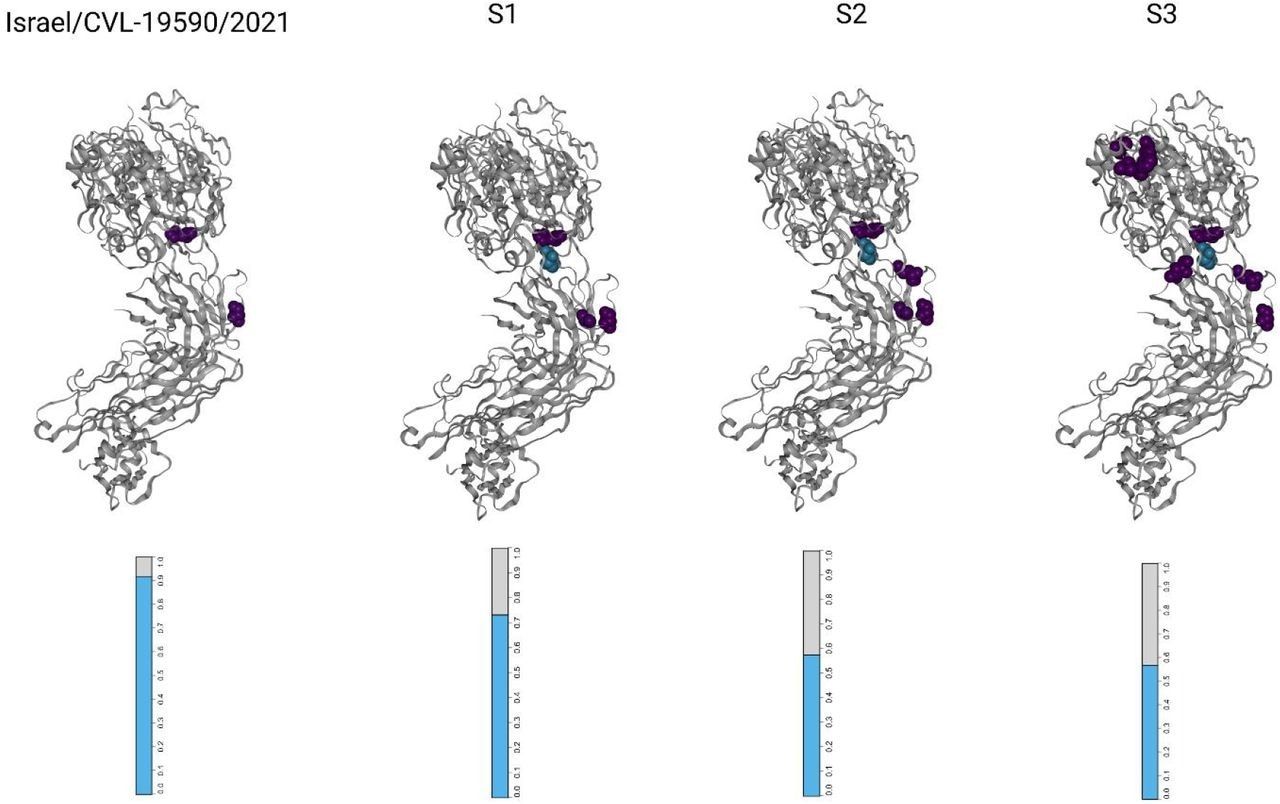
Visualizations of the SARS-CoV-2 receptor binding domains (RBDs), marked according to altered amino acid residues found in each sample. Each mutated residue is colored according to the magnitude of the largest-effect escape mutation at each site, as measured for the REGN10933+REGN10987 monoclonal antibody cocktail (see methods). The bar-plots at the bottom represent the estimated fraction antibodies that were either bound (cyan) or escaped (gray) from neutralization by polyclonal antibodies for each sample (see methods).
The receptor-binding domain (RBD) was the most densely mutated locus, with ~12 mutations per kilobase (mKb), and the Spike gene was the most mutated structural protein. It had 23 unique non-synonymous mutations, eight of which overlapped with the RBD locus. The distribution of mutations in the accessory proteins, open reading frame (ORF)8, ORF10, and ORF3a were 10.9, 8.6, and six mKb, respectively.
It is highly likely that in the first month of infection, the patient's humoral immunity was composed of endogenous antibodies and the REGEN-COV mAbs. Presumably, the mutations acquired before the collection of S1 evaded neutralization by the REGEN-COV mAb cocktail and the endogenous antibodies produced before treatment with Rituximab.
The second REGEN-COV mAb treatment enabled greater escape from antibody binding and decreased the efficacy of the antibody treatment even further. Subsequently, SARS-CoV-2 infectivity assays performed two months after the second round of mAb treatment showed infectious viral shedding. While RBD mutations continued to accumulate for another five months, they showed no correlation with increased antibody escape. Due to the immunosuppressive effects of the Rituximab treatment, the humoral response remained negligible. Overall, even two rounds of REGEN-COV treatments failed to achieve SARS-CoV-2 clearance, and SARS-CoV-2 infection remained unresolved in the patient.
Conclusions
The study demonstrated that SARS-CoV-2 survived two rounds of mAb treatments and remained infectious and viable despite an exceptionally high serum nAb concentration in immunocompromised patients. The adaptive immunity of the patient was severely compromised due to treatment with Rituximab. The mutations in the RBD region of SARS-CoV-2 enabled it to escape from neutralization in the absence of adaptive immunity. Furthermore, emerging mutations disrupted specific immunomodulatory viral functions, providing SARS-CoV-2 selective advantage in the immunocompromised host.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
SARS-CoV-2 evolution and evasion from multiple antibody treatments in a cancer patient, Guy Shapira, Chen Weiner, Reut Sorek Abramovich, Odit Gutwein, Nir Rainy, Patricia Benveniste-Levkovitz, Ezra Gordon, Adina Bar Chaim, Noam Shomron, medRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.25.22276445, https://www.medrxiv.org/content/10.1101/2022.06.25.22276445v1