The early immune response following natural infection shapes the long-term outcome in coronavirus disease 2019 (COVID-19) outpatients. Studies have associated progression to severe COVID-19 with early transcriptomic signatures of retinoic acid-inducible gene I (RIG-I)-induced type I interferon (IFN) signaling and subsequent elevation in the plasma levels of IFN-induced chemokines, such as C-X-C motif chemokine ligand 10 (CXCL10) and CXCL11. Thus, there is an urgent unmet medical need to identify host immune biomarkers predictive of clinical and immunological outcomes in SARS-CoV-2-infected patients.
About the study
In the present study, researchers used 108 samples and data from a clinical trial in SARS-CoV-2-infected outpatients to characterize the trajectory of their immune response at two-time points. First, they determined the correlation between early immune markers and subsequent disease progression, control of viral shedding, and SARS-CoV-2-specific T cell and antibody responses for seven months. Next, they determined and compared these associations in patients who contracted COVID-19 following mRNA vaccination.
The team used proteomics-based machine-learning models for predicting patient outcomes. Further, they validated their findings using data from 54 individuals enrolled in another clinical trial.
They used the ribonucleic acid-sequencing (RNA-seq) and Olink data to characterize each patient's early transcriptomic and proteomic response trajectory. First, deriving the blood transcriptional modules (BTM) from RNA-seq data, they calculated the enrichment score of different immune modules. Next, they combined the enrichment scores and Olink measurements into a single dataset for the trajectory analysis. Furthermore, the team captured the non-linear dynamics of the modules and proteins.
The researchers identified 15 immune modules and ten plasma proteins that varied as a function of time since symptom onset, with a false discovery rate [FDR] of less than 0.05. Of these, 16 immune modules/ proteins showed non-linear dynamics, as indicated by a quadratic regression model.
The researchers used a previously established tool named xCell to characterize the changes in the composition of blood immune cells over time. They noted that the activation of corresponding immune cells rather than the changes in their composition drove the immune trajectories of all 16 immune modules.
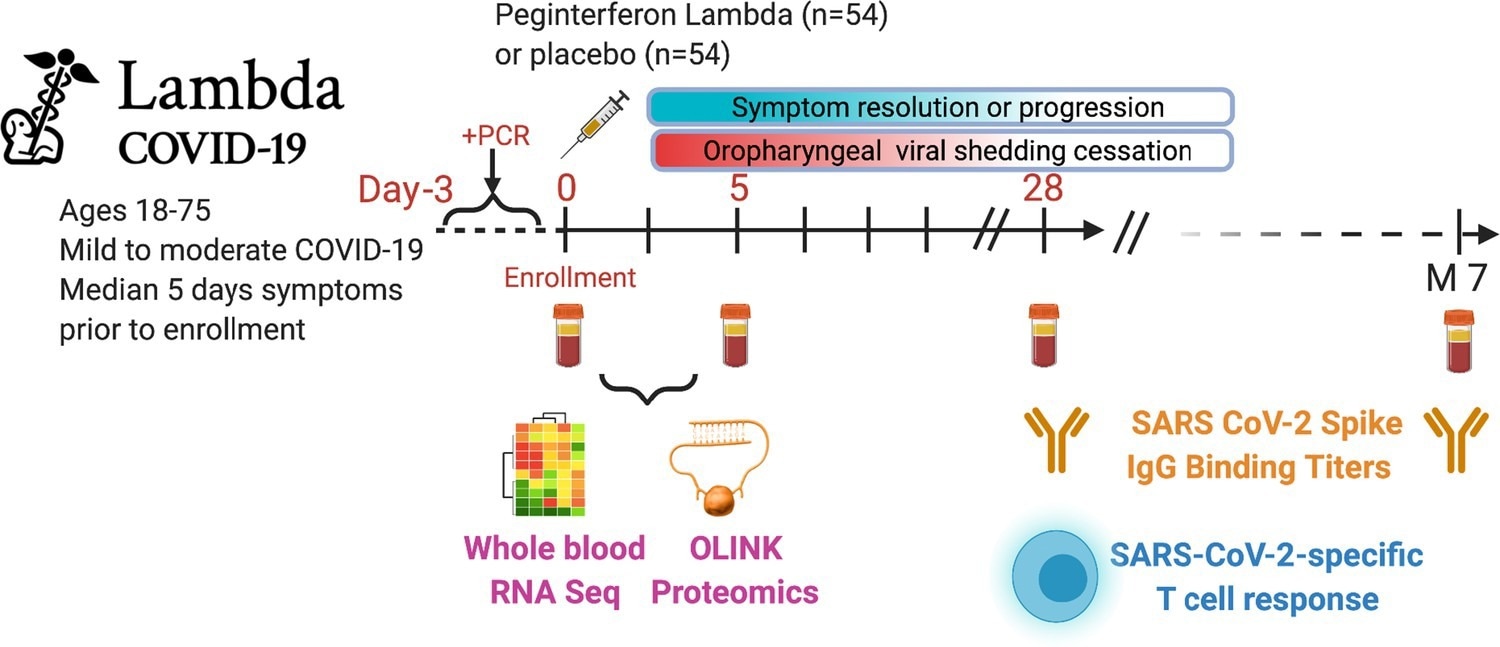 Outpatients (n=108) with PCR-confirmed severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) infection and swab obtained within 72 hr of randomization were enrolled in a phase 2 clinical trial of subcutaneous Peginterferon lambda vs. placebo. In-person follow-up visits were conducted at days 1, 3, 5, 7, 10, 14, 21, 28, and month 7 post-enrollment, with assessment of symptoms and vitals and collection of oropharyngeal swabs for SARS-CoV-2 testing. Blood obtained at days 0 and 5 were evaluated by whole blood transcriptomics (RNA sequencing), plasma proteomics (Olink), and SARS-CoV-2-specific antibodies. Clinical outcomes assessed included duration of symptoms and duration of virological shedding. Immunological outcomes assessed including SARS-CoV-2-specific T cell responses at day 28, and antibody responses at day 28 and month 7. Created with biorender.com.
Outpatients (n=108) with PCR-confirmed severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) infection and swab obtained within 72 hr of randomization were enrolled in a phase 2 clinical trial of subcutaneous Peginterferon lambda vs. placebo. In-person follow-up visits were conducted at days 1, 3, 5, 7, 10, 14, 21, 28, and month 7 post-enrollment, with assessment of symptoms and vitals and collection of oropharyngeal swabs for SARS-CoV-2 testing. Blood obtained at days 0 and 5 were evaluated by whole blood transcriptomics (RNA sequencing), plasma proteomics (Olink), and SARS-CoV-2-specific antibodies. Clinical outcomes assessed included duration of symptoms and duration of virological shedding. Immunological outcomes assessed including SARS-CoV-2-specific T cell responses at day 28, and antibody responses at day 28 and month 7. Created with biorender.com.
Study findings
Transcriptomic and proteomic profiling revealed a sequential activation of IFN signaling, T cells, and B cells within two weeks of COVID-19 symptom onset. Further, the researchers noted an association between RIG-I-triggered early type I IFN plasma levels and multiple COVID-19 outcomes up to seven months. The COVID-19 patient outcomes encompassed COVID-19 progression, SARS-CoV-2 shedding, and the SARS-CoV-2-specific T-cell and antibody responses.
The RIG-I helicase domain interacts with SARS-CoV-2 RNA to induce an inhibitory effect on viral replication. It does so independently of downstream IFN upregulation via mitochondrial antiviral-signaling protein (MAVS). Thus, the study data showed an inverse correlation between plasma RIG-I levels and SARS-CoV-2 load.
In addition, plasma RIG-I levels showed a modest correlation with several intracellular proteins, including deoxyribonucleic acid (DNA) fragmentation factor subunit alpha (DFFA), an intracellular protein involved in cell death. This finding suggested that plasma RIG-I levels reflect an increase in gene expression and cellular apoptosis.
The researchers noted an association between higher expression of three chemotactic receptors (CCR2) ligands, monocyte chemoattractant protein-1 (MCP1), MCP2, and MPC3, with positive immunological outcomes. It reduced viral load in the nasopharynx and increased SARS-CoV-2-specific T cells in the patient's plasma. This finding illustrated that while essential, an overexpression of CCR2 signaling might lead to severe COVID-19 symptoms and tissue damage. Thus, COVID-19 therapeutic strategies must manage the delicate balance between its positive and negative effects.
Conclusions
The machine-learning-based study models accurately predicted all these outcomes using only seven to 10 plasma protein markers, attaining an area under the curve (AUC) of 0.85. Intriguingly, there were stark similarities between early proteomic signatures and adaptive immune responses following natural infection and vaccination.
The Federal Drug Administration had issued emergency use authorization (EUA) for multiple drugs and monoclonal antibodies (e.g., sotrovimab and nirmaltrevir/ritonavir) for COVID-19 treatment. These therapeutics could reduce the risk of hospitalization and mortality in outpatient settings; however, their supplies remain limited.
In such a grim scenario, measuring the plasma proteins during the initial COVID-19 diagnosis could help identify the patients who would most benefit from these therapies. Furthermore, the study model could predict the degree of variation in immune memory following natural infection and vaccination.
Journal reference:
- Zicheng Hu, Kattria van der Ploeg, Saborni Chakraborty, Prabhu S Arunachalam, Diego AM Mori, Karen B Jacobson, Hector Bonilla, Julie Parsonnet, Jason R Andrews, Marisa Holubar, Aruna Subramanian, Chaitan Khosla, Yvonne Maldonado, Haley Hedlin, Lauren de la Parte, Kathleen Press, Maureen Ty, Gene S Tan, Catherine Blish, Saki Takahashi, Isabel Rodriguez-Barraquer, Bryan Greenhouse, Atul J Butte, Upinder Singh, Bali Pulendran, Taia T Wang, Prasanna Jagannathan, Early immune markers of clinical, virological, and immunological outcomes in patients with COVID-19: a multi-omics study, eLife (2022) DOI: https://doi.org/10.7554/eLife.77943, https://elifesciences.org/articles/77943#s3