Background
The human microbiome is a vital source of diagnostic indicators and therapies since it varies between bodily regions and is not isolated. This variability underscores the need for a pan-body approach that considers the microbiota across multiple organ systems, ensuring a more holistic understanding of disease processes. Recent research has emphasized the complicated relationship between non-transferable illnesses and microbes in human tissues, opening up new possibilities for studying disease causation, discovering biomarkers, and generating new treatments.
Studies have found that site-specific microbiome alterations are associated with illness onset and progression. Metagenomic studies like the Human Microbiome Project (HMP), the International Human Microbiome Consortium (IHMC), and the American Gut Project have analyzed thousands of microbial samples. Microbial imbalances have been linked to inflammatory disorders, indicating that the microbiota is crucial to illness development and resolution.
About the study
In the present study, researchers adopted a pan-body, pan-disease approach, examining multiple body sites simultaneously to study microbiome alterations in single-organ disease and multimorbidity. They investigated diagnostic patterns, antimicrobial resistance (AMR), functional capabilities, and potential disease associations and assessed their functional abilities.
From 2021 to 2023, researchers collected samples from individuals suffering from chronic inflammatory conditions of the lungs, eyes, oral cavity, heart, skin, and intestine. They obtained dental plaque, saliva, throat, skin, stool, and eye samples. The samples underwent next-generation and metagenomic sequencing (average sequencing depth, 5.3 gigabases).
Researchers obtained 1,931 high-quality samples from 515 patients, a 3.7-metagenome yield per patient. They excluded samples with inadequate quantity or poor quality of DNA. They analyzed metagenomics related to AMR. Subsequently, researchers performed biosynthetic gene clusters (BGC) prediction. They developed a novel prioritization strategy to identify BGCs with the highest therapeutic potential, based on enrichment or depletion patterns within specific disease cohorts. They investigated whether a BGC exhibited enrichment or depletion within specific disease cohorts. They compared them to the MIBiG database to identify compounds produced by the BGCs.
Researchers generated species genome bins (SGBs) and probed their potential links to diseases. To assess the novelty of the SGBs, they used references like GTDB r214, the Unified Human Gastrointestinal Genome Collection, and the Singapore Platinum Metagenomes Project, including 99,376 genomes. SGBs were known in the case of a match to a known source and novel if there were no matches. Researchers computed the relative microbiome compositions at the genus level and performed a differential abundance analysis at the species level. They computed an embedding based on the MinHash distances for beta diversity to separate the metagenomes by specimen type. Shannon diversity indicated alpha-diversity species richness. Researchers used the Uniform Manifold Approximation and Projection (UMAP) for analysis. Center-log ratios (CLR) denoted normalized abundances for species-cohort-sample combinations. This pan-body approach enabled a more nuanced analysis of microbiome diversity across distinct body sites. They tested dietary data related to the disease context to assess potential confounding.
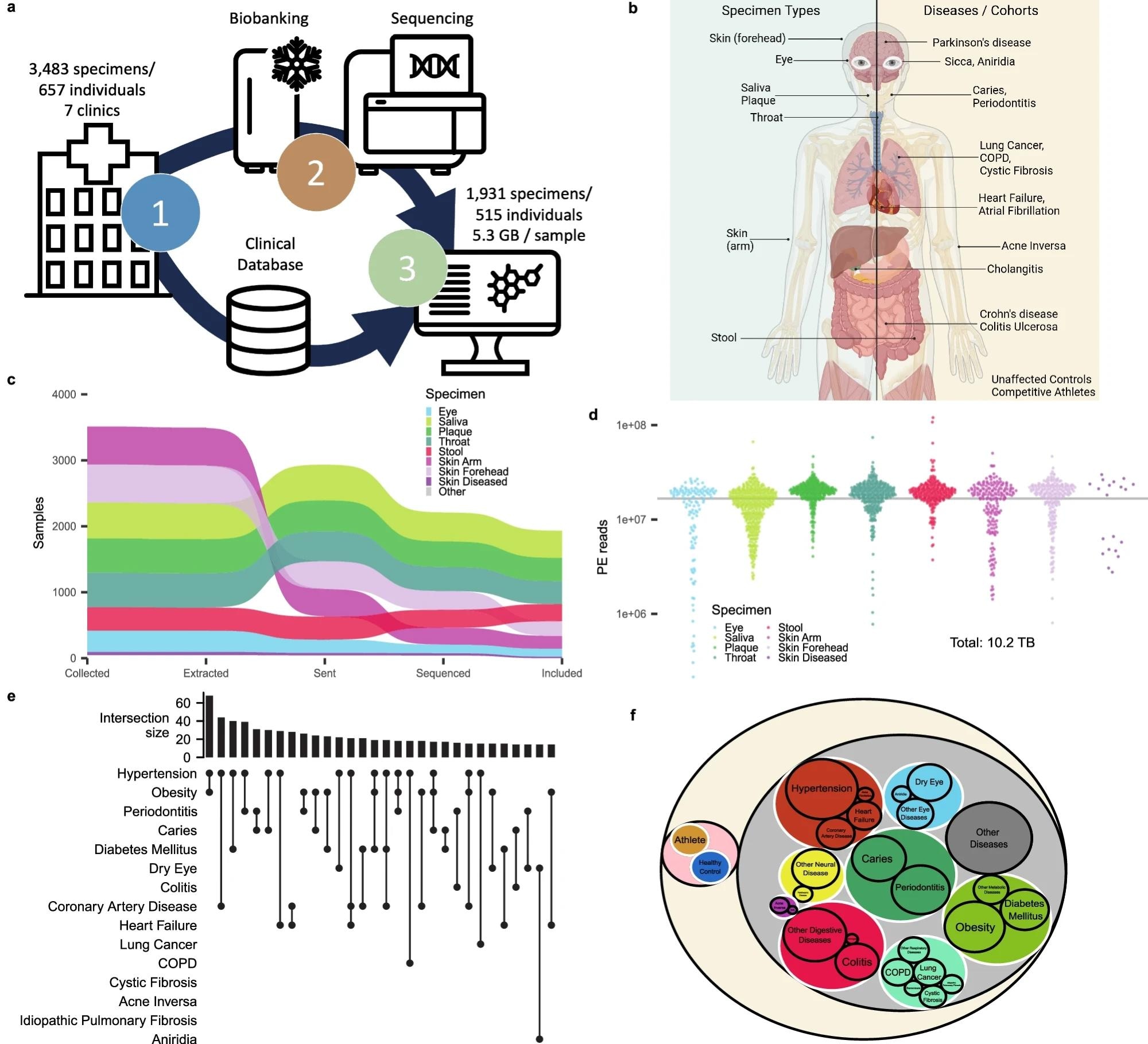 a Schematic Workflow describing the sample (upper arrow) and data flow (lower arrow) between clinicians, microbiology, and data science. The clinical data were kept separated from the measurement of microbiomes and only combined after measurement in the computational analysis. b Clinical sampling was focused on seven biospecimens (left blue part). We included patients from a wide range of clinical diseases, which allowed us to analyze the diagnostic potential of different specimen types across diseases. Created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. c Sankey plot for the number of samples included in the study at different intervals of the data generation process in relation to our quality control strategy. Specimen types are ordered vertically at each step in the pipeline by frequency of the respective specimen. d Number of reads for each sample colored by specimen. The horizontal line represents the 5 gigabase threshold at a paired-end read length of 150 bp. e Pruned upset plot displaying the dataset's most frequent co-occurrence of diseases. The combinations are ordered with decreasing frequency, marking the combination of Hypertension and obesity as the most common comorbidity in our study. f Ontology was used throughout the study to group diseases by biological systems and separate healthy control from diseased patients. Areas are proportional to the number of patients falling into each category. Patients may be represented multiple times if multiple diseases are diagnosed.
a Schematic Workflow describing the sample (upper arrow) and data flow (lower arrow) between clinicians, microbiology, and data science. The clinical data were kept separated from the measurement of microbiomes and only combined after measurement in the computational analysis. b Clinical sampling was focused on seven biospecimens (left blue part). We included patients from a wide range of clinical diseases, which allowed us to analyze the diagnostic potential of different specimen types across diseases. Created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. c Sankey plot for the number of samples included in the study at different intervals of the data generation process in relation to our quality control strategy. Specimen types are ordered vertically at each step in the pipeline by frequency of the respective specimen. d Number of reads for each sample colored by specimen. The horizontal line represents the 5 gigabase threshold at a paired-end read length of 150 bp. e Pruned upset plot displaying the dataset's most frequent co-occurrence of diseases. The combinations are ordered with decreasing frequency, marking the combination of Hypertension and obesity as the most common comorbidity in our study. f Ontology was used throughout the study to group diseases by biological systems and separate healthy control from diseased patients. Areas are proportional to the number of patients falling into each category. Patients may be represented multiple times if multiple diseases are diagnosed.
Results
The study showed significant variations in microbial abundances across specimen types and diseases. The team identified 583 unexplored SGBs, 189 of which were significantly associated with disease. Previously undescribed and annotated SGBs harbored 28,315 probable BGCs, with 1,050 significant associations with diseases. Oral microbiome specimens (saliva and plaque) exhibited more undescribed genomes, accounting for 72% of novelty in SGBs. These findings highlight the value of investigating multiple specimen types simultaneously, as certain body sites, like the oral cavity, demonstrated significantly higher novelty in microbial genomes. Patients with comorbidities present higher alterations in the human microbiome, regardless of the body site.
Importantly, the study underscores that comorbidities amplify microbiome disruptions, leading to more pronounced shifts in microbial diversity. Patients with multiple chronic conditions had consistently higher microbiome alterations across all body sites studied, indicating that multimorbidity is a key factor in microbiome variation.
Corynebacterium pseudogenitalium and Staphylococcus epidermidis showed a significantly higher abundance in the skin of coronary heart disease patients. The team detected enrichment of Capnocytophaga gingivalis in salivary samples obtained from individuals with aniridia, Lachnoanaerobaculum saburreum species in plaque samples from those with aniridia, and Porphyromonas endodontalis in interdental plaque samples provided by obese individuals. Bacteroides cellulosilyticus abundance was significantly lower in fecal samples of individuals with digestive diseases. Streptococcus vestibularis was considerably less abundant in salivary samples of individuals with aniridia and more numerous in those from individuals with Parkinson’s disease.
Across all samples, the mef(A) gene that encodes resistance against macrolide antimicrobials was the most prevalent. The most observed genes conferring resistance against carbapenem were New Delhi metallo beta-lactamases (NDM) and oxacillin-hydrolyzing (OXA) carbapenemases. The findings indicated that patients with resistance genes on the skin carried the same resistance genes in the gut.
Researchers found 11 diminished microbial abundance in vegetarian stool, with species like Alistipes inops, Phascolarctobacterium faecium, and Bifidobacterium more abundant among omnivorous participants. Dialister CAG 357 strain, related to inflammation, showed higher levels among omnivores. Only one diet-related hit, Saccharimonas sp013333645, derived from dental plaque, remained. The findings suggest confounding factors influence metagenomic patterns, but disease signals persist.
The team’s novel approach to BGC prioritization, which involves evaluating BGC enrichment or depletion patterns, revealed several BGCs with significant therapeutic potential. Specifically, the study identified BGCs producing compounds with high similarity to known antibiotics, highlighting their relevance for further in vitro exploration and drug development.
The study highlights using pan-body pan-disease microbiomics to diagnose and manage diseases. The findings call for future research on the functionality of identified BGCs, considering factors like similarity, biotechnological suitability, and quality measures. Further exploration in vitro could lead to medical discoveries, including antibiotic compounds.
Journal reference:
- Schmartz, G.P., Rehner, J., Gund, M.P. et al. Decoding the diagnostic and therapeutic potential of microbiota using pan-body pan-disease microbiomics. Nat Commun 15, 8261 (2024), DOI: 10.1038/s41467-024-52598-7, https://www.nature.com/articles/s41467-024-52598-7