Ion mobility separation is a highly robust method used for separating isomeric or isobaric compounds. Particularly in tandem with high-resolution accurate mass spectrometry, ion mobility separation significantly improves substance identification and characterization capabilities, and offers an extra physical property of compounds — the Collisional Cross Section (CCS) values.
In this case, Bruker applies Trapped Ion Mobility Spectrometry (TIMS) on the new timsTOFTM instrument to various model compounds, such as lipids or isomeric trisaccharides.
| Keywords |
Instrumentation and Software |
| Isomeric trisaccharides |
timsTOF |
| Lipids |
Data Analysis 5.0 |
| CCS values |
|
| imeXTM technology |
|
In real-world samples, isomeric compounds are frequently hard to identify when depending exclusively on LC-MS techniques, because chromatographic separation is not always adequate to acquire clean MS/MS spectra. timsTOF solves this challenge with its exceptional ion mobility expansion technology (imeXTM) that delivers adjustable ion mobility resolution for suitable separation of isomeric compounds, both in LC experiments or with direct infusion.
Methods
Combinations of isomeric analytes were prepared from the pure standard compounds (melezitose, raffinose, and maltotriose (Sigma-Aldrich) as well as 18:1 (Δ9-cis) PC, 18:1 (Δ6-cis) PC, 16:0-18:0 PC, 16:0-18:1 PC, and 16:0-18:2 PC (Avanti Polar Lipids)). Next, ImeX technology was used to modify the mobility resolution according to the unique TIMS working principle (see Figure 1).
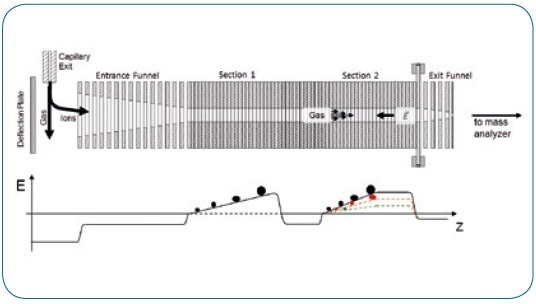
Figure. 1 Schematic of TIMS functionality.
Sample mixtures were examined by direct infusion with and without sequential MS/MS fragmentation. For the calibration of CCS values and mass accuracy, ESI low concentration Tunemix was used (Agilent Technologies).
Results
timsTOF with imeX technology enabled effective separation of homologous or isomeric compounds by altering the ion mobility resolution to the prerequisite level. ImeX technology simplified the selection of the ideal ion mobility resolution mode. While imeX Survey mode is most appropriate for mobility separation of the complete mass range, imeX Detect is desired for establishing collisional cross sections. Maximum ion mobility resolution is realized in imeX Ultra mode for highest separation efficiency (Figure 2).
![imeX technology provides three resolution modes demonstrated for the isomers raffinose, melezitose, and maltotriose: C18H32O16 (1 μg/mL, m/z 527.1583 as [M+Na]+).](https://www.news-medical.net/image-handler/picture/2019/3/Job_6039_Art10_Pic2.jpg)
Figure 2. imeX technology provides three resolution modes demonstrated for the isomers raffinose, melezitose, and maltotriose: C18H32O16 (1 μg/mL, m/z 527.1583 as [M+Na]+).
A number of compound mixtures were examined concerning mass accuracy and their isotopic pattern quality in addition to MS/MS fragmentation patterns after ion mobility separation (see Figure 3).
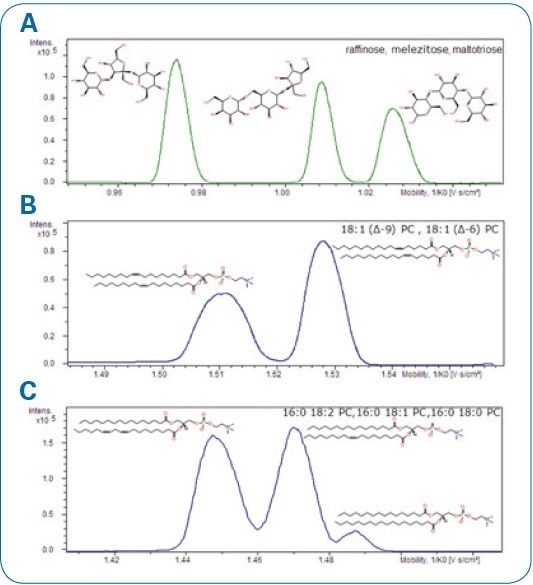
Figure. 3 Mobilograms display the separation of selected compounds. A. Raffinose, melezitose, and maltotriose. B. 18:1 (Δ6-cis)-PC and 18:1 (Δ9-cis)-PC lipids. C.16:0-18:2 PC, 16:0-18:1 PC, and 16:0-18:0 PC lipids.
Bruker accomplished quasi-baseline separation for a series of homologous lipids that enables acquisition of pure MS/MS spectra from the precursor ions that no longer overlap. Mass accuracy was in the ppm range and with superior isotopic pattern quality, as can be seen in Figure. 4.
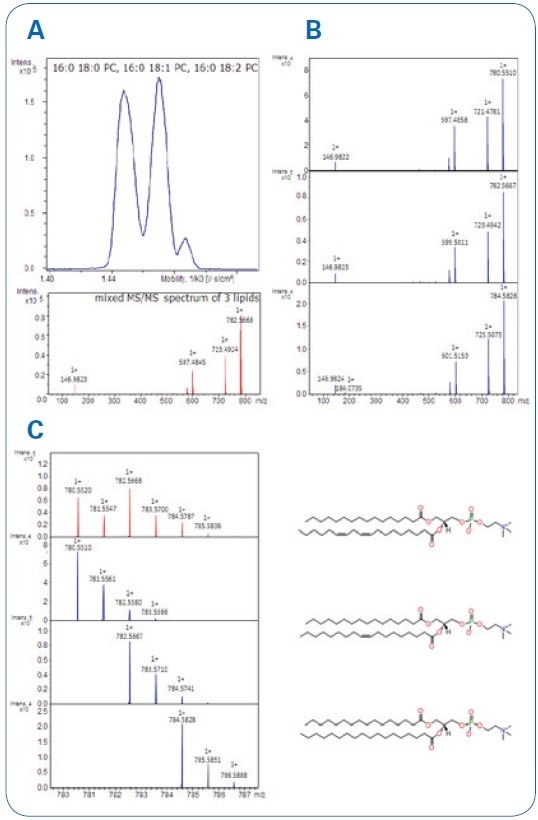
Figure 4. A Mobility separation of the three lipids with mixed MS/MS spectrum (acquisition without IM separation and a full isotope pattern isolation). B Clean MS/MS spectra after IM-separation. C Clean iso-topic patterns showing as well mass accuracies and isotopic pattern qualities (mSigma; value ranges from 0-1000; <10 is excellent).
CCS values were established with excellent precision (RSD <0.5%, n=20) for selected compounds shown in Table 1. As the CCS value is a physical property of the ion, it offers an extra level of confidence to a library-based unknown identification workflow.
Table 1. CCS values of selected compounds (N2)
| Compound |
1/K0 [Vscm- 2] |
CCS value (Å2) |
% RSD |
| Raffinose |
1.021 |
209.818 |
0.1 |
| Melezitose |
0.984 |
202.214 |
0.2 |
| Maltotriose |
1.040 |
213.722 |
0.2 |
| 18:1 (Δ6-cis) PC |
1.520 |
309.608 |
0.1 |
| 18:1 (Δ9-cis) PC (DOPC) |
1.498 |
305.127 |
0.2 |
| 16:0-18:0 PC |
1.476 |
300.800 |
0.1 |
| 16:0-18:1 PC |
1.460 |
297.553 |
0.2 |
| 16:0-18:2 PC |
1.443 |
294.101 |
0.2 |
Summary
High-resolution ion mobility separation used on oligosaccharide isomer and lipid homolog characterization expose the benefits of this technology, for instance, accurate CCS values, accurate mass and true isotopic patterns, and ion mobility purified MS/MS spectra.
Conclusions
- timsTOF instrument offers >200 RP in the ion mobility dimension and provides exact CCS values
- imeX technology permits adjustment of the ion mobility resolution for various purposes
- High-resolution ion mobility enables separation of isobaric precursor ions and attainment of clean MS/MS spectra
Acknowledgements
Produced from materials originally authored by Sven W. Meyer; Peter Sander; Annette Michalski; Detlev Suckau from Bruker Life Sciences Mass Spectrometry, Bremen, Germany
About Bruker Life Sciences Mass Spectrometry

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.