Samples in shotgun proteomics frequently present analytical difficulties, since they are often greatly complex and thousands of peptides can easily be detected in a narrow mass range.
As a result, peptide separation is often carried out by nano-flow UHPLC, which offers a number of benefits, such as high peak capacity and ESI compatible solvent composition and flow rate.
In spite of the growing rapidity and sensitivity of mass spectrometers, improving their capacity to investigate a greater number of ions eluting from the column at any one time, enhanced chromatographic separation still offers an immense improvement in the number of proteins identified.
Additionally, highly reproducible peak areas and retention times are vital for quantitative analyses of greater complexity. To address each of these demands, a reliable HPLC system is critical.
This study focused on the assessment of the four critical chromatographic performance indicators of the Bruker nanoElute: retention time variation, peak area variation, peak width and peak capacity, in order to test the nanoElute for the tough demands of advanced proteomic analytics.
Experimental
Samples
Aliquots of a commercially available tryptic digest of HeLa cells were diluted with 0.1% formic acid in water to a concentration of 100 ng/µL. As can be seen in the below table, for chromatographic separation, a curved gradient with three varied lengths was used.
The gradient is made up of solvent A, 0.1% formic acid in water and solvent B, 0.1% formic acid in acetonitrile, with the temperature of the separation column preserved at 50 °C and a flow rate of 400 nL/min.
The 90 minute gradient made four technical replicates, while the 120 minute and 240 minute gradients each made five. Each of these was measured using a setup both with and without a trap column. Swapping between setups with and without a trap column is a simple and convenient task with the software.
An Impact II QTOF mass spectrometer furnished with a CaptiveSpray nanoBooster Source with pure ACN was employed for MS detection. The apparatus was used in ESI positive mode, gaining full scan MS and MS/ MS data with the InstantExpertise™ routine.
This is a self-adapting auto MS/MS process developed to gather the best quality results, regardless of the sample’s complexity and concentration. It employs a sophisticated parent ion selection process, joined with a variable MS/MS acquisition rate [1] and comes already installed in the impact II instrument control software (otofControl).
Data Analysis
Bruker’s Compass DataAnalysis was employed to analyze the chromatographic data. Eight peptides, uniformly dispersed in time over the gradient, were chosen from the HeLa chromatogram. EICs were created for these peptide masses, and comparisons of the peak area, retention time and peak widths were carried out between the technical replicates.
| Liquid chromatography |
| Instrument: |
Bruker nanoElute™ |
Gradient Conditions: |
90 min Gradient |
120 min Gradient |
240 min Gradient |
Composition B |
| Column: |
Acclaim PepMap™ RSLC; 75 µm x 50 cm |
0 min |
0 min |
0 min |
2% B |
| Mobile phase A: |
Water, 0.1% formic acid |
60 min |
90 min |
180 min |
15% B |
| Mobile phase B: |
ACN, 0.1% formic acid |
90 min |
120 min |
240 min |
25% B |
| Trap column loading: |
100% mobile phase A |
100 min |
130 min |
250 min |
35% B |
| Flow rate: |
400 nL/min |
110 min |
140 min |
260 min |
95% B |
| Injection volume: |
2 µL |
120 min |
150 min |
270 min |
95% B |
| Column oven: |
50 °C |
|
|
|
|
|
| Mass Spectrometry |
| Instrument: |
Bruker impact II QTOF mass spectrometer |
| Ion source: |
CaptiveSpray nanoBooster in positive ion mode |
| Capillary: |
1600 V |
| Dry Gas: |
3.0 L/min |
| Dry Temperature: |
150 °C |
| nanoBooster: |
0.20 Bar |
Results and Discussion
Generally, the gradient length in shotgun proteomics fluctuates between 90 minutes to 240 minutes. With the combination of the pressure limit of the new nanoElute of 1000 bar and a column oven, it is possible to achieve 50 cm column lengths.
The nanoElute Method Editor enables the generation of any gradient with just a few mouse clicks, and all usual wash and equilibration stages are included automatically.
Performance
For any quantitative study, highly reproducible chromatographic separation is vital. It is essential that the LC delivers an eluent flow, the gradient mixture and the injection volume exactly to guarantee the ability to reproduce each chromatographic test.
Comparisons were made between a number of technical replicates with a 90 minute, 120 minute and 240 minute gradient, and the most significant performance criteria were identified in order to assess the performance of the instrument.
As can be seen in Figure 2, regardless of the setup used, base peak chromatograms were steady over all gradient lengths, with a time shift occurring only as a result of the greater dead volume when a trap column was employed. Both column configurations produced high retention time stability and peak area reproducibility (Figure 3).
Even with the lengthy 240 minute gradient, retention time drifts of just a few seconds were attained. The retention time variation was under 0.5% for all gradient lengths.
In addition, the peak area reproducibility was outstanding, with variations below 10% for the majority of the comparisons and ≤5% for every experiment in which a trap column was employed (Table 1).
Predictably, the 90 minute gradient offered the slenderest peaks for both column setups, 11 s full width half maximum (FWHM) (Figure 4). The FWHM values grew correspondingly alongside the gradient’s length, leading to a peak capacity between 500 and 700 (Figure 5).
This guarantees the greatest separation to allow the mass spectrometer to study as many precursors as possible.
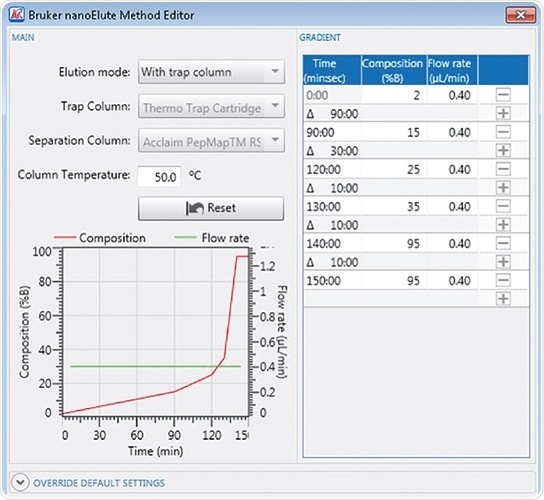
Figure 1. Bruker nanoElute Method Editor
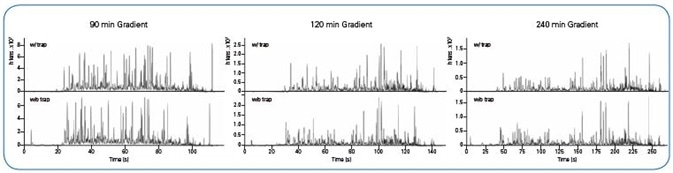
Figure 2. Representative Base Peak Chromatograms for all gradient lengths and column setups. Chromatograms in the upper row are obtained by using a trap column and in the lower row without a trap column.
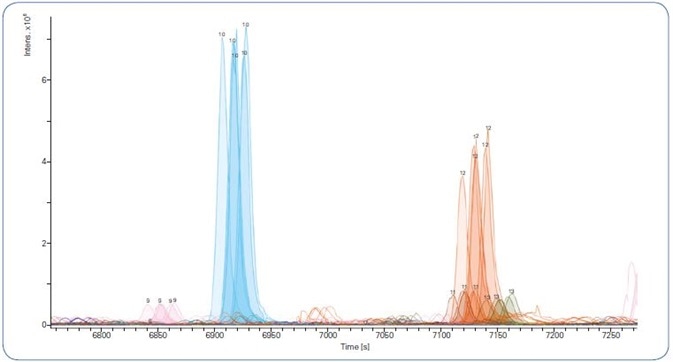
Figure 3. Extracted Ion Chromatograms showing retention time reproducibility of selected peptides across 5 technical replicates using a 120 min gradient.
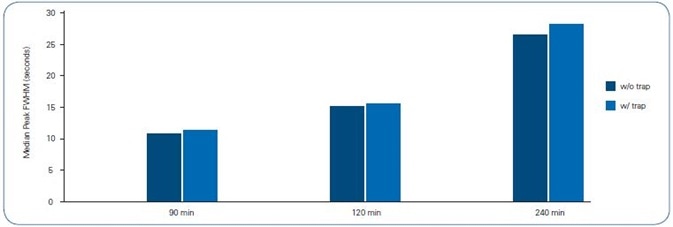
Figure 4. Median Peak FWHM of 8 peptides across each gradient
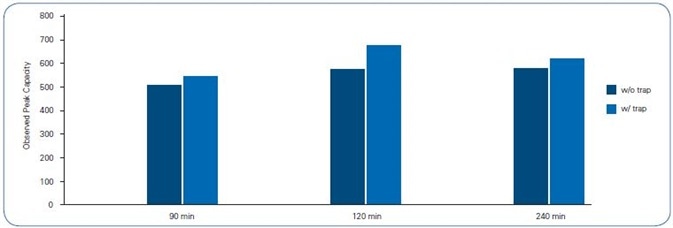
Figure 5. Peak Capacity of 12 peptides across each gradient
Table 1. Summery of chromatographic performance values
| . |
| Median Retention Time Variation |
| 90 min |
120 min |
240 min |
| w/o trap |
w/ trap |
w/o trap |
w/ trap |
w/o trap |
w/ trap |
| 0.16% |
0.05% |
0.18% |
0.35% |
0.11% |
0.26% |
| Median Area Variation |
| 90 min |
120 min |
240 min |
| w/o trap |
w/ trap |
w/o trap |
w/ trap |
w/o trap |
w/ trap |
| 13% |
4% |
8% |
5% |
3% |
4% |
| Median Peak FWHM (seconds) |
| 90 min |
120 min |
240 min |
| w/o trap |
w/ trap |
w/o trap |
w/ trap |
w/o trap |
w/ trap |
| 11 |
11 |
15 |
16 |
26 |
28 |
| Observed Peak Capacity |
| 90 min |
120 min |
240 min |
| w/o trap |
w/ trap |
w/o trap |
w/ trap |
w/o trap |
w/ trap |
| 514 |
546 |
577 |
676 |
581 |
620 |
Conclusion
For carrying out high performance proteomics analyses, the nanoElute is the ideal partner to the impact II. In boasting both excellent performance and strong reliability, it outstrips other cutting edge nano-UHPLC systems.

Altogether, the nanoElute delivers simple-to-operate method configuration and advanced diagnostic processes with just one press. It offers:
- High retention time stability
- High Peak Area reproducibility
- Narrow peaks
- High peak capacity
- Simple gradient configuration
- Automatic diagnostic tool
- Trap column switchable with a push of a button
References
[1] Bruker App Note LC-MS 81
Introducing New Proteomics Acquisition Strategies with the compact™ – Towards the Universal Proteomics Acquisition Method
About Bruker Life Sciences Mass Spectrometry

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.