Synthetic cannabinoids are usually screened by analyzing urine since it is a noninvasive matrix. None or just a small concentration of the parent cannabinoid will be left for detection because of the metabolism in the human body. Therefore, both the parent compound and drug metabolites need to be screened.
Hydroxylation is one common metabolism for AKB-48. The hydroxylation can occur on varied carbon positions leading to different structural isomers. The broad range of several metabolites and the introduction of novel drug analogs make urine screening an extremely difficult task that needs the flexibility of detecting new drug metabolites as well as high confidence for known substances.
In order to address this challenge, a mass spectrometer with the potential to deliver exact mass measurements, clear MS/MS spectra, and high confidence in isotopic pattern distribution is the first step to gain an understanding of a complex sample. Additionally, the potential to detect and differentiate structural isomers can be improved further by using the ion-mobility-separation offered by the timsTOF.
Methods
As AKB-48 metabolites were not commercially available, these metabolites were prepared in-house[1] and used for measurements. Next, two mixtures of different AKB-48 hydroxylation metabolites were prepared with a concentration of 125 ng/mL each. Before the ion mobility separation process, chromatographic separation was used. Detection was carried out on an ion mobility separation-time-of-flight instrument (timsTOF, Bruker Life Sciences Mass Spectrometry).
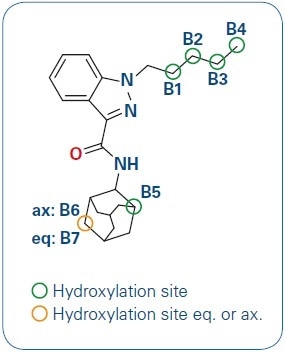
Figure 1. AKB-48 and its hydroxylation sites.
Instrumentation
| . |
| UPLC |
Ultimate 3000 Rapid Separations LC, Thermo Scientific |
| Column |
HSS T3, 2.1 x 150 mm, 1.8 μm, Waters |
| Temperature |
60 °C |
| Mobile phase |
A = H2O, 10 mM Ammonium formate and 0.05% formic acid
B = ACN, 0.05% formic acid |
| Gradient |
Multistep gradient from 57%-93% B in 8 min |
| Flow rate |
0.5 mL/min |
| Injection |
1 μL |
| MS |
timsTOF mass spectrometer, Bruker Life Sciences Mass Spectrometry |
| Scan mode |
Full scan TOF MS with IMS in Ultra mode |
| Ionization |
ESI +2500 V |
| MS-Calibration |
sodium formate cluster |
| IMS-Calibration |
tuning mix, low concentration |
Results and Discussion
In the initial step, focus was given to the LC separation of the metabolites to allow a clear characterization of each metabolite (see Figure 2). The metabolites were ionizing as a mixture of [M+Na]+ and [M+H]+ adducts, both showing exceptional isotope pattern distributions and mass accuracies (Figure 3 as one representative example), and thus demonstrating the high data quality for a first identification process.
![Extracted Ion Chromatograms for [C23H31N3O2 +H]+ with m/ z = 382.2489 ± 3 mDa. Two different mixtures 1 and 2 of hydroxylated AKB-48 metabolites were prepared and measured.](https://www.news-medical.net/image-handler/picture/2019/3/Job_6041_Art3_Pic2.jpg)
Figure 2. Extracted Ion Chromatograms for [C23H31N3O2 +H]+ with m/ z = 382.2489 ± 3 mDa. Two different mixtures 1 and 2 of hydroxylated AKB-48 metabolites were prepared and measured.
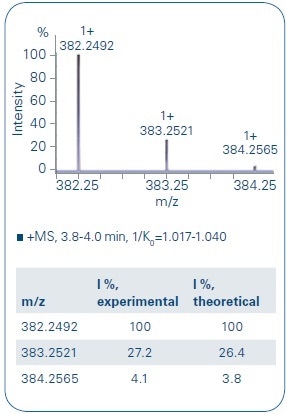
Figure 3. Comparison of the experimental data of B4 and the theoretical isotope pattern distribution shows a good overlay. The monoisotopic signal as well as the following isotopes show only a small deviation to the theoretical values (mass accuracy: - 0.8 ppm, mSigma: 4.9)
In a subsequent step, Extracted Ion Mobilagram (EIM) traces of different adduct species ([M+Na]+ and [M+H]+) were investigated. Both types of adducts revealed a clear separation on the ion mobility axis. Sodium adducts showed a higher value around 1.04–1.08 [V∙s/cm2] (see Figure 4, orange), while the protonated species showed signals of the inverse mobility 1/K0 [V∙s/cm2] with values of approximately 1.00–1.04 (see Figure 4, blue).
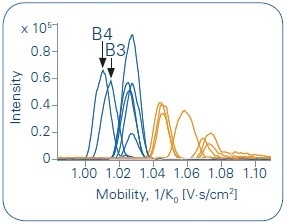
Figure 4. Overlay of all Extracted Ion Mobilograms (EIM) of the protonated species (in blue, m/z = 382.2492) and the sodium adducts (in orange, m/z = 404.2312). A grouping of the different species is clearly given.
Compared to the other metabolites, the inverse mobility is shifted to lower 1/K0 values for the metabolites obtained from hydroxylation at the end of the alkyl chain (B3 and B4).
As shown in Figure 5, the sodium adducts can be used to achieve a clear differentiation of metabolites derived from hydroxylations at the adamantyl moiety. These metabolites (B5-B7) demonstrate higher 1/K0 values than those showing a hydroxylation at the alkyl chain (B1-B4). Fascinatingly, unlike the other metabolites, B1 does not fit into the grouping of the other isomers. It shows a 1/K0 value in the middle of adamantyl-moiety and alkyl-chain.
![Display of EIM traces of the different metabolites as [M+Na]+. In orange B2-B4 are displayed referring to metabolites showing hydroxylation of the alkyl chain. In green B5-B7 are displayed referring to hydroxylation on the adamantyl moiety. B1 in blue is an exception as it shows 1/K0 values between those of the two groups.](https://www.news-medical.net/image-handler/picture/2019/3/Job_6041_Art3_Pic5.jpg)
Figure 5. Display of EIM traces of the different metabolites as [M+Na]+. In orange B2-B4 are displayed referring to metabolites showing hydroxylation of the alkyl chain. In green B5-B7 are displayed referring to hydroxylation on the adamantyl moiety. B1 in blue is an exception as it shows 1/K0 values between those of the two groups.
After human liver microsome incubations (HLM incubation), two of the major metabolites are B7 and B4[2]. B4 and B7 are co-eluting based on the used LC separation technique. Thus, the differentiation of both compounds is not distinctly given through LC-MS only techniques. A clear differentiation of both compounds is achieved by adding IMS separation. Both the [M+Na]+ and the [M+H]+ EIM trace reveal a baseline separation of both isomeric structures (Figure 6).
Along with the isotope pattern distribution, the exact mass, and verification using fragmentation, CCS values serve as an extra parameter for identification and validation of possible metabolites of designer drugs.
![Overlay of EIM traces for B4 and B7. A baseline separation for both adduct types ([M+H]+ and [M+Na]+) is given.](https://www.news-medical.net/image-handler/picture/2019/3/Job_6041_Art3_Pic6.jpg)
Figure 6. Overlay of EIM traces for B4 and B7. A baseline separation for both adduct types ([M+H]+ and [M+Na]+) is given.
Conclusions
- Metabolites showing a hydroxylation on the alkyl chain or a hydroxylation at the adamantyl group can evidently be differentiated based on the mobility of the sodium adducts
- Ion Mobility together with the timsTOF provides an extra separation technique for differentiating between different AKB-48 metabolites in tandem with excellent values in isotopic pattern distribution and mass accuracy
- A clear grouping of the CCS values is provided based on the region of hydroxylation
References
[1] Jakob Wallgren, Svante Vikingsson, Anders Johansson, Martin Josefsson, Henrik Green, Johan Dahlén, Xiongyu Wua, Peter Konradsson, Synthesis and identification of an important metabolite of AKB-48 with a secondary hydroxyl group on the adamantyl ring, 2017, Tetrahedron Letters, (58), 1456-1458.
[2] Svante Vikingsson, Martin Josefsson and Henrik Green, Identification of AKB-48 and 5F-AKB-48 Metabolites in Authentic Human Urine Samples Using Human Liver Microsomes and Time of Flight Mass Spectrometry, 2015, Journal of Analytical Toxicology, (39), 6, 426-435.
About Bruker Life Sciences Mass Spectrometry

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.