Researchers can work with low sample input and higher numbers of samples due to the miniaturization and scale-down of reagent and reaction volumes for low-input RNA-Seq workflows. The accurate delivery of reagent volumes emerges as a vital step to maintain technical reproducibility and data quality as the reaction volumes are decreased.
In this article results from miniaturization of the Clontech SMART-Seq® v4 Ultra® Low Input RNA Kit for Sequencing and Illumina Nextera® XT workflows are presented for the production of high-quality RNA-Seq samples utilizing a compact desktop microfluidic dispensing technology.
Materials and methods
SMART-Seq v4 ultra low input RNA kit
SMART® technology (Figure 1) is based on non-templated nucleotides which are added by an MMLV-based reverse transcriptase (RT) during cDNA synthesis when it reaches the 5’ end of the mRNA.
Next, template switching happens when a specially designed SMART oligo bearing a complementary sequence to these non-templated nucleotides hybridizes to the first-strand cDNA. The RT switches to utilizing the SMART oligo for further cDNA synthesis from utilizing the mRNA as a template.
This ensures that the 5’ end of the mRNA is captured, and for simpler amplification and enrichment of full-length cDNA, enables specific sequences to be added to each end of the cDNA.
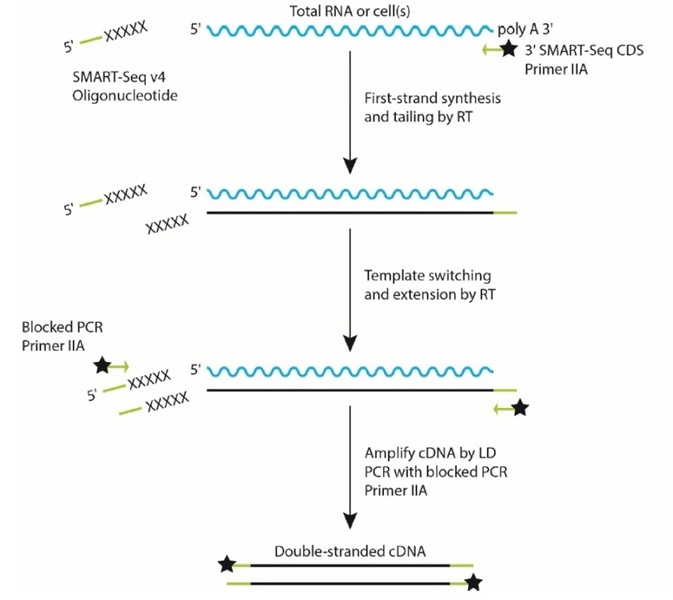
Figure 1. SMART technology for cDNA synthesis. Image Credit: FORMULATRIX
Mantis low volume dispensing
The MANTIS® is a low volume, easy to program, non-contact, low dead-volume, reagent dispenser. The Mantis has a dispense range of 100 nl – 2 ml, may be configured with up to 24 different reagents, can dispense any volume into any well, and is compatible with any plate density up to 1536.
Reagents can plug directly into microfluidic chips to minimize dead volume. By using pipette tips as reagent reservoir, dead volumes can be decreased further to 6 μl s. This feature is perfect for dispensing NGS sample prep reagents.

Figure 2. MANTIS® microfluidic dispensing system. Image Credit: FORMULATRIX
cDNA synthesis and library prep
Using the standard full volume reaction conditions, 10 ng aliquots of mouse brain total RNA and aliquots of 1000 cells (K562 cell line) and negative control samples were processed and compared with samples that were processed by utilizing reduced scale miniaturized reaction volumes using SMART-Seq v4 Ultra Low Input RNA Kit.
Reagent volumes can be observed in Table 1. In order to amplify the cDNA products, 8 cycles of PCR were utilized. The resulting cDNA products were characterized by utilizing the Fragment Analyzer (AATI) and quantified via PicoGreen® assay
Table 1. cDNA Synthesis reagent volumes for full scale and miniaturized reactions. Buffers, primers and Master Mix reagents were dispensed using the MANTIS low-volume chip. Source: FORMULATRIX
| Reagent |
Standard Reaction (µL) |
Miniaturized Reaction (µL) |
| Sample |
1 |
1 |
| 10x Reaction Buffer |
9.5 |
1.6 |
| 3’ SMART CDS Primer |
2 |
0.5 |
| SMART Master Mix |
7.5 |
1.9 |
| PCR Master Mix |
30 |
7.5 |
| 10x Lysis Buffer |
1 |
0.3 |
| AMPure® XP beads |
50 |
12.8 |
| Elution Buffer |
17 |
17 |
Illumina sequencing libraries were prepared from 100 pg aliquots of the amplified cDNA products via the Nextera XT fragment library kit (Illumina) utilizing reduced scale reaction volumes, as shown in Table 2. For illustrative purposes, a comparison to standard full reaction volumes is included.
Table 2. Nextera XT Fragment library prep reagent volumes. Reagents were dispensing using the MANTIS® low-volume chip. Source: FORMULATRIX
| Reagent |
Standard Reaction (µL) |
Miniaturized Reaction (µL) |
| Sample |
5 |
1.25 |
| Tagment DNA Buffer |
10 |
2.5 |
| Amplicon Tagment Mix |
5 |
1.25 |
| Neutralize Tagment Buffer |
5 |
1.25 |
| Nextera PCR Master Mix |
15 |
3.75 |
| Index 1 |
5 |
1.25 |
| Index 2 |
5 |
1.25 |
| AMPure XP Beads |
30 |
7.5 |
| Resuspension Buffer |
50 |
12.5 |
For both the full scale and miniaturized cDNA synthesis samples, miniaturized Nextera XT reaction conditions were employed. To amplify the sequencing libraries 12 cycles of PCR were utilized.
In order to process decreasing input of cells (K562), miniaturized cDNA synthesis and library prep conditions were utilized with samples of 1000, 100, 10, 1 cell using 8, 11, 14, 17 cycles of PCR respectively, to amplify the cDNA products from the decreasing amount of cells in each sample.
Sequencing and data analysis
Indexed samples were pooled and sequenced by utilizing MiSeq with 2x75 bp reads. Using CLC, reads were trimmed by CLC Genomics Workbench (ver. 8.5.1) and mapped to rRNA and the mitochondrial genome.
Next, unmapped reads were mapped with CLC to the human genome (hg19) and mouse genome (mm10) with RefSeq masking, to generate uniquely mapped reads and % reads which mapped to RefSeq annotations, including introns, exons, and intergenic regions.
The number of genes identified in each library was established by the number of genes with a RPKM ≥0.1. Expression levels of different genes were gathered by utilizing CLC Genomics Workbench after mapping to RefSeq.
Results
cDNA synthesis
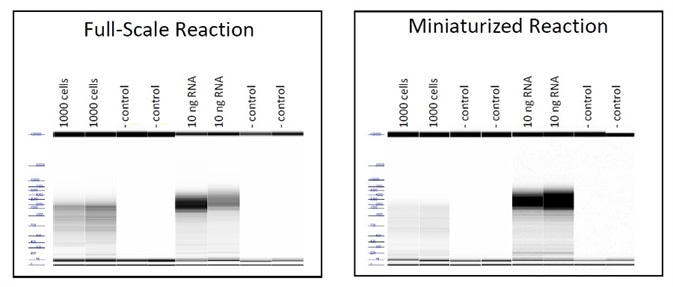
Figure 3. Electropherograms demonstrating equivalent size distribution of cDNA products for full and small scale reactions. Image Credit: FORMULATRIX
Table 3. cDNA product yield (ng/μl) measured with Pico-Green assay. Source: FORMULATRIX
| |
Average Yield (n=6) |
| 1000 cells - full reaction |
4.63 +/- 0.88 |
| 1000 cells - miniaturized reaction |
4.26 +/- 0.96 |
| 10 ng - full reaction |
5.42 +/- 0.58 |
| 10 ng - miniaturized reaction |
4.88 +/- 0.54 |
Sequencing library prep
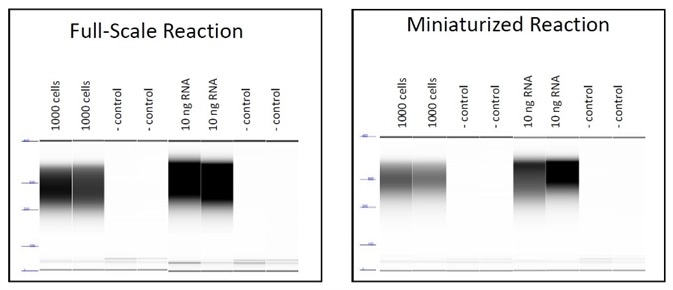
Figure 4. Electropherograms of final fragment libraries demonstrating equivalent size distribution for full and small scale cDNA synthesis reactions for both cell and RNA input samples. Image Credit: FORMULATRIX
Sequencing results – comparison of full-scale and miniaturized reaction
Table 4. RNA-Seq data analysis comparing full-scale and miniaturized reaction conditions. Source: FORMULATRIX
| |
Full Reaction |
Mini Reaction |
Full Reaction |
Mini Reaction |
| Input |
10 ng total RNA |
1000 cells |
| # of reads |
903962 |
982866 |
484780 |
484740 |
| Mapped to rNA |
3.6% |
3.0% |
1.7% |
0.2% |
| Mapped to RefSeq |
92% |
85% |
90% |
90% |
| Mapped uniquely to RefSeq |
86% |
80% |
80% |
79% |
| Mapped to exons |
78% |
77% |
83% |
84% |
| Mapped to introns |
17% |
18% |
13% |
12% |
| # of transcripts with at least 0.1 RPKM |
16651 |
16615 |
12550 |
10285 |
| Pearson Correlation |
0.98 |
0.94 |
Table 5. cDNA product yield (ng/Âμl) measured with Pico-Green assay. Source: FORMULATRIX
| # K562 Cells |
# PCR Cycles (cDNA amplification) |
Average Yield (n=6) |
| 1000 |
8 |
5.13 +/- 0.67 |
| 100 |
11 |
3.77 +/- 0.99 |
| 10 |
14 |
5.05 +/- 1.83 |
| 1 |
17 |
3.38 +/- 0.91 |
Table 6. RNA-Seq data analysis from decreasing number of cells using miniaturized sample prep conditions. Source: FORMULATRIX
| |
1000 cells (n=6) |
100 cells (n=6) |
10 cells (n=6) |
1 cell (n=6) |
| # of reads |
3.4 M
+/- 0.7 M |
2.2 M
+/- 0.4 M |
2.4 M
+/- 0.4 M |
0.8 M
+/- 0.2 M |
| Mapped to rRNA |
0.21%
+/- 0.06% |
0.21%
+/- 0.09% |
0.14%
+/- 0.07% |
0.1%
+/- 0.03% |
| Mapped to RefSeq |
87%
+/- 3% |
91%
+/- 2% |
89%
+/- 2% |
89%
+/- 3% |
| Mapped uniquely to RefSeq |
78%
+/- 3% |
83%
+/- 2% |
82%
+/- 2% |
82%
+/- 3% |
| Mapped to exons |
85%
+/- 8% |
89%
+/- 1% |
91%
+/- 1% |
92%
+/- 1% |
| Mapped to introns |
11%
+/- 1% |
8%
+/- 1% |
7%
+/- 1% |
6%
+/- 1% |
| # of transcripts with at least 0.1 RPKM |
15855
+/- 449 |
16092
+/- 575 |
15682
+/- 247 |
10171
+/- 1127 |
Pearson Colleration
(to full scale reaction) |
0.96
+/- 0.02 |
0.95
+/- 0.01 |
0.92
+/- 0.01 |
0.82+/-
0.03 |
Conclusions
When utilizing the Formulatrix Mantis microfluidic dispensing system, reducing reagent volumes for cDNA synthesis with the Clontech SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing generates results which are comparable to full-scale reaction volumes.
The average yield of amplified cDNA from the scaled-down reaction volume workflow is compatible with input samples of cells and total RNA. RNA-Seq data analysis metrics are comparable between the full-scale and miniaturized reaction conditions.
The combination of high-yield cDNA synthesis chemistry and high-precision low-volume reagent dispensing supplies a workflow which is ideal for large-scale RNA-Seq studies.
References
- Nextera XT DNA Library Prep Protocol Guide (Illumina)
- SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing User Manual (Clontech)
- MANTIS® Operators Guide (Formulatrix)
About FORMULATRIX
FORMULATRIX® was established in 2002 to provide protein crystallization automation solutions. Since then, we've started developing other laboratory automation solutions including the next generation of liquid handlers using microfluidic technology.
Headquartered in Bedford, Massachusetts, we supply software and robotic automation solutions to leading pharmaceutical companies and academic research institutions around the world. Our team works tirelessly to provide the best products in the industry with support that is second to none.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.