Sponsored Content by BellBrook LabsReviewed by Olivia FrostMay 10 2024
Cyclic GMP-AMP synthase (cGAS) functions as a foreign DNA sensor, triggering an immune response to pathogens by activating the STING (stimulator of interferon genes) receptor. Shortly after its discovery in 2013, abnormal activation of cGAS by self-DNA was found to cause debilitating and often deadly autoimmune disorders, such as systemic lupus erythematosus (SLE) and Aicardi–Goutieres Syndrome (AGS).
Knockout experiments in animal models have demonstrated that blocking cGAS is a possible strategy for therapeutic intervention. This study created Transcreener®-based assays for cGAMP detection to support HTS efforts targeting cGAS. These are homogenous, competitive immunoassays based on direct cGAMP detection with fluorescence polarization (FP) and time-resolved Förster resonance energy transfer (TR-FRET) signals. The assay uses an antibody that is highly specific to cGAMP in the presence of excess ATP and GTP, as well as far-red tracers to measure activity. The direct detection format simplifies the protocol (no coupling enzyme needed) and reduces compound inteference.
Both assays were validated with full-length human cGAS, and pilot screens were performed for cGAS inhibitors using the FP assay. Like other Transcreener assays, the cGAMP assays exhibit the performance qualities required for small-molecule screening, such as sensitivity, robustness (high dynamic range, minimum signal change), and low levels of compound interference.
Developing reliable, HTS-compatible assays for detecting cGAS enzyme activity will boost efforts targeting the cGAS-STING pathway for autoimmune disorders.
The cGAS-STING Pathway Activates the Immune System in Response to Cytosolic DNA
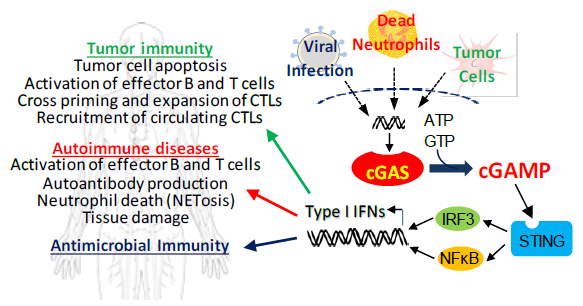
Figure 1. IFN-driven immune responses triggered by cGAS/STING are critical for protection against many types of microbial pathogens and for tumor cell-specific T cell responses in cancer, but activation by self-nucleic acids can contribute to serious autoimmune diseases such as lupus. Image Credit: BellBrook Labs
Mix-and-Read HTS Enzymatic Assay for cGAS Based on Immunodetection of cGAMP
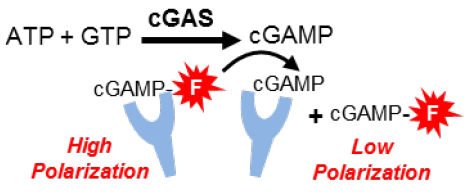
Figure 2. Transcreener cGAS Assay principle: in the competitive fluorescence polarization (FP) immunoassay for cGAMP, enzymatically generated cGAMP displaces a fluorescent tracer from mAb, causing a decrease in its fluorescence polarization. Image Credit: BellBrook Labs
Nanomolar sensitivity and outstanding selectivity
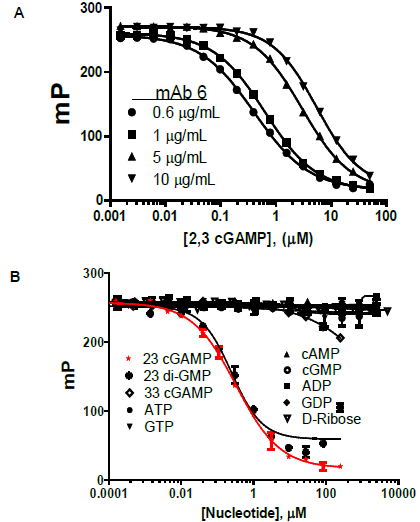
Figure 3. A. Competition curves indicate displacement of tracer by cGAMP and show dependence of dynamic range on mAb concentration. B Specificity of mAb: Competition binding curves show outstanding selectivity for cGAMP vs. cGAS substrates, ATP and GTP, as well as related molecules. Image Credit: BellBrook Labs
Validation for Detection of cGAS Under Initial Velocity
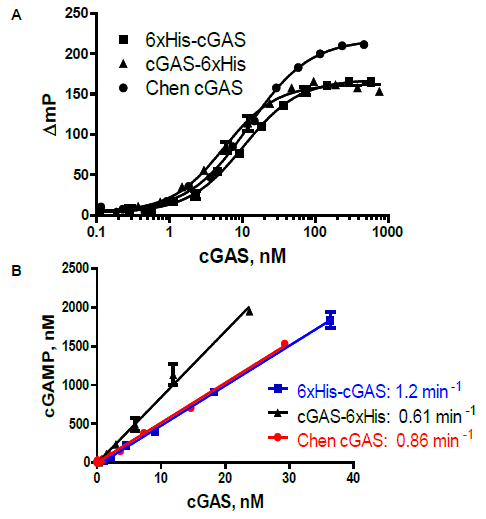
Figure 4. A. Detection of purified, full-length human cGAS. cGAS enzyme reactions contained 100 µM ATP and GTP, 62.5 nM 45 bp ISDna, 60 min reactions. N- and C-terminal His-tagged cGAS was produced at BBL; 6xHis-cGAS was also generously supplied by Z. Chen (UTSW Medical Center). B. Linear response: Polarization data from A. was converted to cGAMP using a standard curve. Image Credit: BellBrook Labs
Substrate and DNA Dependence
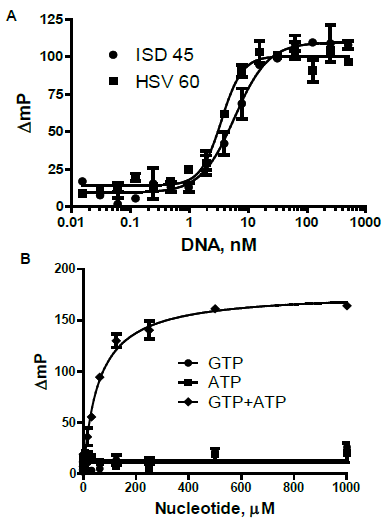
Figure 5. A. DNA dependence cGAS reactions as in Figure 4, with cGAS-6xHis at 10 nM; half maximal responses of 3.5 and 5.9 nM for HSV 60 and ISDna 45, respectively. B. ATP and GTP dependence: ATP and GTP were titrated separately and simultaneously; cGAS reactions as in Figure 4, with cGAS-6xHis at 10 nM. Image Credit: BellBrook Labs
Pilot Screens for cGAS Inhibitors
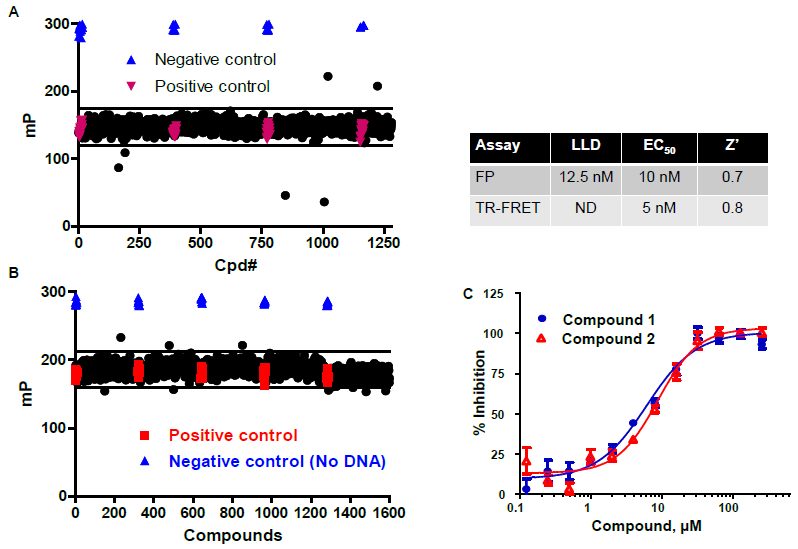
Figure 6. A. Interference screen: The LOPAC library (1280 compounds) was used at 10 µM final concentration. Wells contained all cGAS enzyme reaction components except the cGAS enzyme; positive controls contained 1 µM cGAMP, negative controls contained no cGAMP. B. Pilot screen with 1600 diversity compounds (Life Chemicals): cGAS was used at 10 nM, compounds were at 10 µM; 60 min reaction; negative controls lacked dsDNA (required for cGAS activation); Z = 0.62, Z’ = 0.7. C. Dose response for cGAS inhibition by two hits from pilot screen shown in B. Image Credit: BellBrook Labs
Conclusions
- Competitive immunoassays for cGAMP with FP and TR-FRET readouts were developed, allowing useful detection of cGAMP concentrations, ranging from 0.1 to 50 µM.
- The assays can detect cGAS at low nanomolar concentrations with Z’ values of 0.7 or higher.
- Pilot screens demonstrated acceptable levels of compound interference (< 0.4%) and verified the assay for identifying cGAS inhibitors.
- The Transcreener cGAS Assays will speed up efforts to test small molecule cGAS modulators for the treatment of autoimmune disorders, such as SLE and AGS.
About BellBrook Labs
BellBrook Labs is dedicated to providing scientists with enabling screening tools to accelerate the discovery of more effective therapies. Leveraging its two base platforms, BellBrook has developed easy-to-use assays for hundreds of drug targets.
Transcreener® Biochemical Assay Technology
The proprietary Transcreener HTS Platform uses a highly specific antibody and far-red tracer for fluorescent immunodetection of nucleotides, including ADP, UDP, GDP, AMP, and GMP. Because its based on detection of nucleotides, the assay is universal for use with virtually any enzyme that produces these nucleotides, such as kinases, glycosyltransferases, GTPases, helicases, ATPases, nucleotidases, exonucleases and PDEs. The assay boasts direct detection of many of these enzyme targets (no coupling enzyme needed), simplyifing the protocol and reducing compound interference.
AptaFluor® Biochemical Assay Technology
AptaFluor leverages a spit aptamer technology to directly detect SAH, the common product of Methyltransferases. As the most sensitive HTS methyltransferase activity assay available, AptaFluor dramatically reduces enzyme usage and allows the assay to be run at or below Km for SAM.
Enzolution™ Assay Systems
Enzolution Assay Systems used with Transcreener Assay technology make for a comprehensive assay solution. Enzolution includes the enzyme, substrate, assay plates and buffers required to produce the enzyme reaction. Using these together simplifies researchers' assay needs without the need to spend time and money sourcing enzymes and developing assays.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.