Sponsored Content by BellBrook LabsReviewed by Olivia FrostMay 14 2024
RNA helicases are a group of enzymes that bind to and alter RNA in an ATP-dependent way. They play significant roles in RNA metabolism, gene expression regulation, and innate immunity. Different RNA helicases, particularly those in the dead box helicase (DDX) family, are dysregulated in cancer; however, there have been very few attempts to produce small molecule inhibitors.
High-throughput biochemical assays were assembled for RNA helicases DDX3, DDX5, DD17, RIG-I, and MDA5 using the Transcreener ADP2 Assay. The assay measures RNA-dependent ATPase activity using a highly specific antibody and fluorescent tracer to directly detect ADP produced by the helicases. The assays were developed to accelerate the discovery of selective helicase inhibitors, particularly with helicases that encourage tumor immunity.
Purified enzymes determined the RNA substrate, cofactor requirements, and kinetic parameters. The assays were then optimized for initial velocity detection of RNA-dependent ATPase activity with a Z’ value greater than 0.7, guaranteeing an adequate signal window for inhibitor screening and dose-response assays. Each of the helicase assays were then validated by screening a library of bioactives (Tocris 2.0) and perfoming follow-up dose response measurements.
These assays will allow screening and hit-to-lead/SAR for DDX helicases that drive tumorigenesis and selectivity profiling to prevent off-target effects with RLR helicases that enhance tumor immunity.
Transcreener ADP2 Assay: ADP Detection in a Homogenous Format with a FP, FI, or TR-FRET Readout
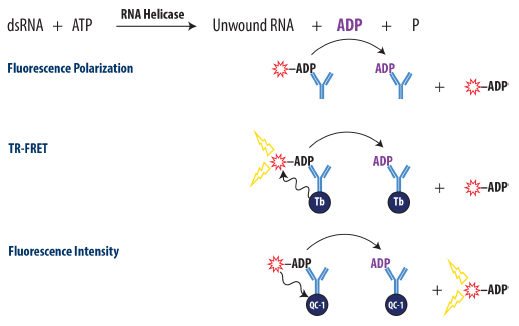
Figure 1. Schematic Overview of the Transcreener ADP2 FP, FI, and TR-FRET Assay. ADP produced by the target enzyme displaces a tracer from the ADP2 antibody, resulting in decreased fluorescence polarization for FP assay, decreased TR-FRET for TR-FRET assay, and increased fluorescent intensity for FI assay. Image Credit: BellBrook Labs
RNA Substrate Optimization
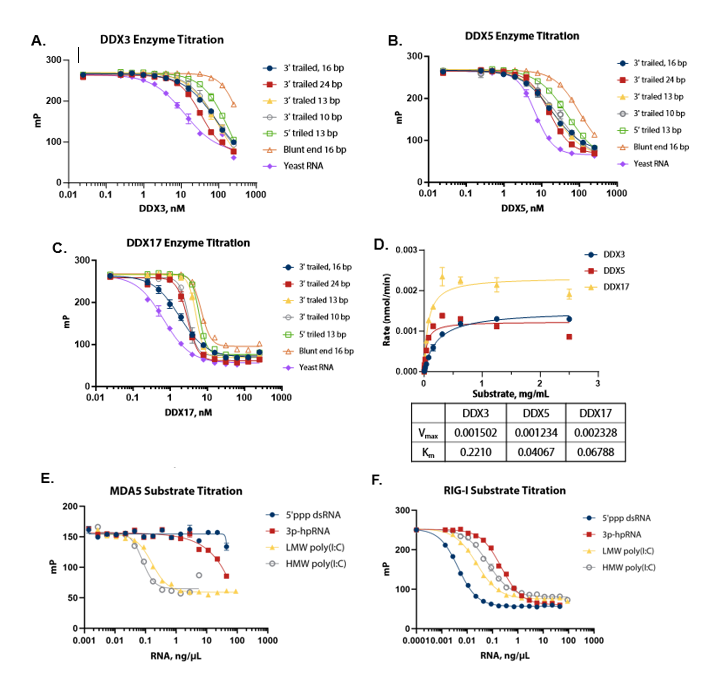
Figure 2. Substrate optimization and Km determination. (A) (B) (C) DDX3, 5, and 17 were titrated with different RNA substrates; yeast RNA was selected as a suitable common substrate for all three enzymes. (D) Michaelis-Menten plot shows the Km value of yeast RNA for different DDX enzymes; a concentration of 1 mg/mL, which is saturating for all enzymes, was selected for standard assay conditions. (E) (F) Different RNA substrates were titrated with MDA5 and RIG-I. HMW poly (I:C) at a concentration of 2 ng/μL was selected for MDA5 and 5’ppp dsRNA at a concentration of 0.2 ng/μL was selected for RIG-I. Image Credit: BellBrook Labs
Detection of RNA-Dependent ATPase Activity
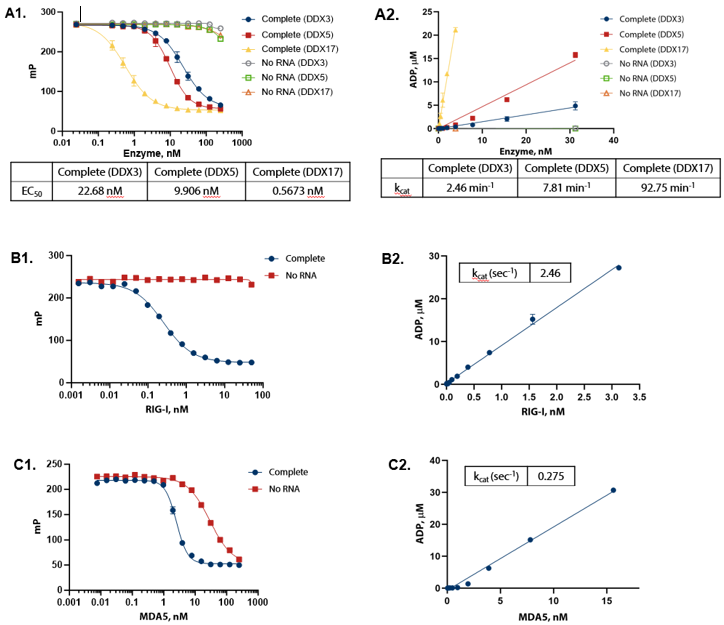
Figure 3. ADP formation shows linear response to enzyme concentration and strict dependence on RNA. (A1) DDX enzymes were titrated in the presence of 100 µM ATP and 1 mg/mL of yeast RNA in Enzyme Assay Buffer D (50 mM Tris (pH 7.5), 2 mM MgCl2 , 0.01% Triton) at 30 °C for one hour. (A2) Conversion of raw data to ADP using standard curves demonstrates that ADP formation is linear with enzyme. (B1) RIG-I (full length) was titrated in the presence of 100 µM ATP (sub-Km concentration) and 0.2 ng/μL 5’ppp dsRNA in Enzyme Assay Buffer C (50 mM Tris (pH 7.5), 50 mM NaCl, 2.5 mM MgCl2 , 0.01% Brij) with 5 mM DTT at 30°C for one hour. (B2) Conversion of raw data to ADP using standard curves demonstrates that ADP formation is linear with enzyme. (C1) MDA5 (AA 299-1025) was titrated in the presence of 100 μM ATP (sub-Km concentration) and 10 ng/μL Poly(I:C) dsRNA in Enzyme Assay Buffer C (50 mM Tris (pH 7.5), 50 mM NaCl, 2.5 mM MgCl2 , 0.01% Brij) with 5 mM DTT and 1.25 mM MnCl2 at 30 °C for two hours. (C2) Conversion of raw data to ADP using standard curves demonstrates that ADP formation is linear with enzyme. Image Credit: BellBrook Labs
Pilot Screen: RIG-I and MDA5
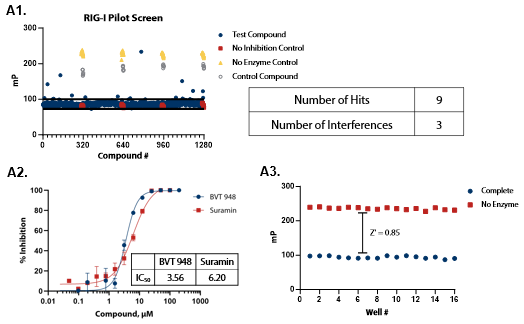
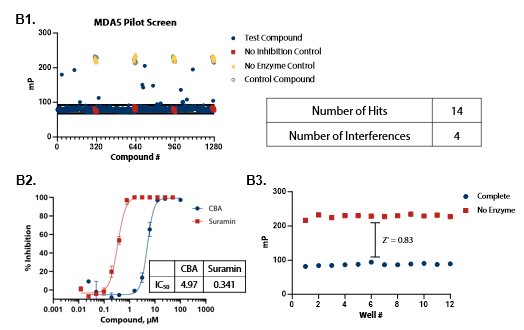
Figure 4. Pilot screens validate the ATPase assay for discovery of novel RIG-I and MDA5 inhibitors. (A1) 1280 compounds from the Tocris 2.0 Library set were screened against RIG-I at 0.9 nM. An interference screen was performed to eliminate compounds interfering with detection reagents. (A2) A selected hit from the pilot screen (BVT 948) and a control inhibitor (Suramin) were tested in dose-response mode. (A3) Complete enzymatic reactions under initial velocity conditions (0.9 nM RIG-I) and ‘no enzyme’ control reactions were plotted to calculate Z’ of 0.85, indicating a robust assay amenable to HTS. (B1) 1280 compounds from the Tocris 2.0 Library were screened against MDA5 (4 nM). (B2) A selected hit from the pilot screen (CBA) and the control compound (Suramin) were tested in dose-response mode. (B3) Z’ determination for MDA5 as in (A3). Image Credit: BellBrook Labs
Pilot Screen: DDX3, DDX5 and DDX17
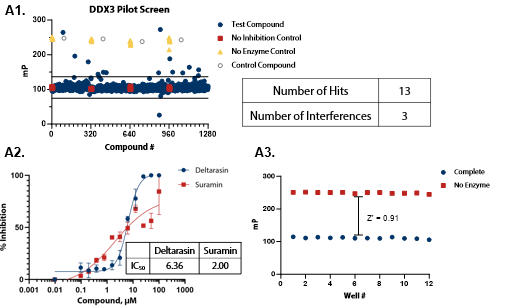
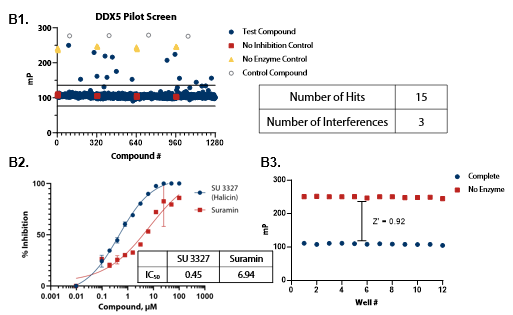
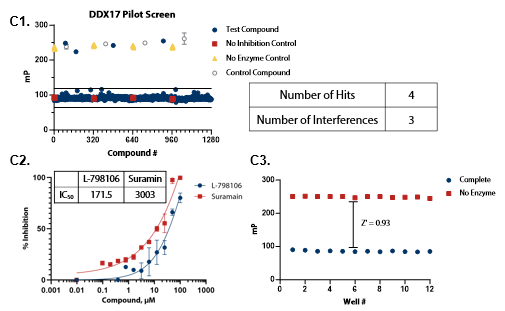
Figure 5. Pilot screens validate the ATPase assay for discovery of novel DDX inhibitors (A1) 1280 compounds from the Tocris 2.0 Library set were screened against DDX3 at 50 nM . (A2) A selected hit from the pilot screen (Deltarasin) and a control inhibitor (Suramin) were tested in dose-response mode. (A3) Complete enzymatic reactions under initial velocity conditions (50 nM DDX3) and ‘no enzyme’ control reactions were plotted to calculate Z’. (B1) DDX5 screen (15 nM). (B2) A selected hit from the pilot screen (SU 3327) and the control compound (Suramin) were tested in dose-response mode with IC50 of 0.45 μM and 6.94 μM, respectively. (B3) Z’ determination for DDX5 as in (A3). (C1) DDX17 screen (2.5 nM). (C2) A selected hit from the pilot screen (L-798106) and the control compound (Suramin) were tested in dose-response mode. (C3) Z’ determination for DDX17 as in (A3). Image Credit: BellBrook Labs
Conclusions
- The Transcreener ADP2 Assay allows for direct, homogenous identification of RNA-dependent ADP production by multiple DDXs, RIG-I, and MDA5 with FP, FI, and TR-FRET readouts.
- The three assay formats (FP, FI, and TR-FRET) exhibit comparable rates of ADP production by all five RNA helicase enzymes at initial velocity settings, demonstrating consistency among the multiple readout formats.
- The assay offers high data quality (Z’ > 0.8) and signal window, delivering a robust HTS assay for various DDXs, RIG-I, and MDA5.
- Pilot screens verified the assay for discovering various DDXs, RIG-I, and MDA5 inhibitors and quantifying the IC50 values.
- The Transcreener ADP2 Assay can help identify biologically pertinent inhibitors for many DDXs, RIG-I, and MDA5, and similar ADP-producing enzymes.
About BellBrook Labs
BellBrook Labs is dedicated to providing scientists with enabling screening tools to accelerate the discovery of more effective therapies. Leveraging its two base platforms, BellBrook has developed easy-to-use assays for hundreds of drug targets.
Transcreener® Biochemical Assay Technology
The proprietary Transcreener HTS Platform uses a highly specific antibody and far-red tracer for fluorescent immunodetection of nucleotides, including ADP, UDP, GDP, AMP, and GMP. Because its based on detection of nucleotides, the assay is universal for use with virtually any enzyme that produces these nucleotides, such as kinases, glycosyltransferases, GTPases, helicases, ATPases, nucleotidases, exonucleases and PDEs. The assay boasts direct detection of many of these enzyme targets (no coupling enzyme needed), simplyifing the protocol and reducing compound interference.
AptaFluor® Biochemical Assay Technology
AptaFluor leverages a spit aptamer technology to directly detect SAH, the common product of Methyltransferases. As the most sensitive HTS methyltransferase activity assay available, AptaFluor dramatically reduces enzyme usage and allows the assay to be run at or below Km for SAM.
Enzolution™ Assay Systems
Enzolution Assay Systems used with Transcreener Assay technology make for a comprehensive assay kit. Enzolution includes the enzyme, substrate, assay plates and buffers required to produce the enzyme reaction. Using these together simplifies researchers' assay needs without the need to spend time and money sourcing enzymes and developing assays.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.