Sponsored Content by BellBrook LabsReviewed by Olivia FrostMay 10 2024
Three Prime Repair Exonuclease 1 (TREX1) is an essential component of the innate immune response. It is the primary exonuclease responsible for the degradation of cytosolic DNA. The reduction of cytosolic DNA inhibits the cGAS/STING Pathway, which is required for intrinsic anti-tumor immunity.
Consequently, TREX1 plays a pro-tumorigenic role in cancer, and inhibiting its activity is a promising method for future monotherapy and combination therapies. TREX1 Inhibitors are currently being investigated, although there are no peer-reviewed studies with quantitative data on individual components.
This study developed a robust, HTS-compatible assay technique for screening small-molecule TREX1 modulators and for selectivity profiling them.
In this approach, TREX1 cleaves Interferon Stimulatory DNA (ISDna) to create dAMP, which is subsequently detected by the Transcreener dAMP Exonuclease Assay, a far-red, competitive fluorescence polarization (FP) assay.
This article shows how the Transcreener dAMP Exonuclease Assay can offer a robust and dependable method for discovering TREX1 inhibitors. It shows the assay's selectivity and sensitivity, followed by its ability to achieve robust assay signals (>100 mP) with less than 250 pM of TREX1 and a Z’ Value of 0.88.
The assay was verified with a pilot screen of 3,056 pharmacologically active compounds, which revealed numerous inhibitors. The assay enables simple triaging of non-stoichiometric inhibitors and profiling selectivity against comparable targets.
The Transcreener dAMP Exonuclease Assay from BellBrook Labs accelerates the discovery of TREX1 antagonists, offering potential molecules that can be used for new cancer treatments.
Transcreener dAMP Exonuclease Assay: Measure TREX1 Enzyme Activity with a Mix-and-Read FP Readout
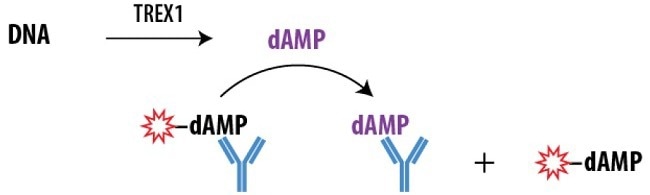
Figure 1. Schematic Overview of the Transcreener dAMP FP Assay. The Transcreener dAMP Detection Mixture contains a dAMP AlexaFluor® 633 tracer bound to a dAMP antibody. dAMP produced by TREX1 displaces the tracer, which rotates freely, causing a decrease in the FP observed. Image Credit: BellBrook Labs
Robust HTS-ready assay procedure
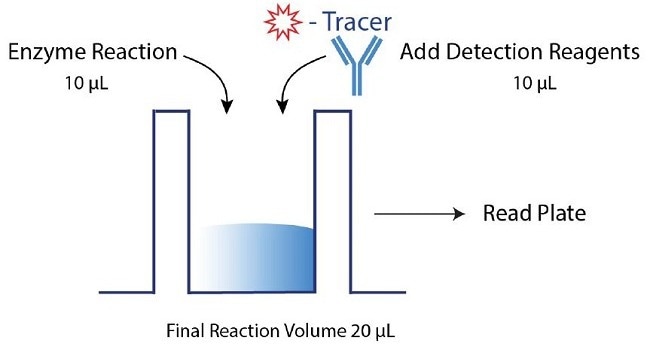
Figure 2. Transcreener dAMP FP Assay Procedure. The Transcreener dAMP Exonuclease Assay relies on a simple, but robust, mix-and-read procedure that is compatible with 96, 384, and 1536-well formats. The assay is performed by running an enzyme reaction followed by the addition of detection reagents. Data can then be obtained with a compatible plate reader. Here, a 10 µL enzyme reaction was completed, followed by adding 10 µL detection reagents in a 384-well format. Image Credit: BellBrook Labs
Nanomolar sensitivity and outstanding selectivity
A.

B.

Figure 3. Assay Sensitivity and Specificity of dAMP Antibody. A. Competition curves indicate displacement of tracer by dAMP and show dependence of antibody concentration on the dynamic range. Detection limit up to 1 nM can be achieved by using 5 µg/mL Ab. B. Competition curves show outstanding selectivity for dAMP vs. dCMP, dGMP, and related molecules. Image Credit: BellBrook Labs
Detection of TREX1 under initial velocity

Figure 4. TREX1 Enzyme Titration and Kinetics. A. TREX1-dependent production of dAMP. B. Conversion of mP to dAMP using a standard curve demonstrates that product formation is linear with enzyme. C. Continuous detection of dAMP formation by TREX1 showing linearity over time until substrate DNA is depleted (10X enzyme, 2.5 nM), indicating detection under initial velocity conditions (350 nM ISDna and 250 pM TREX1 for 60 min). Image Credit: BellBrook Labs
DNA dependence and Z’ determination
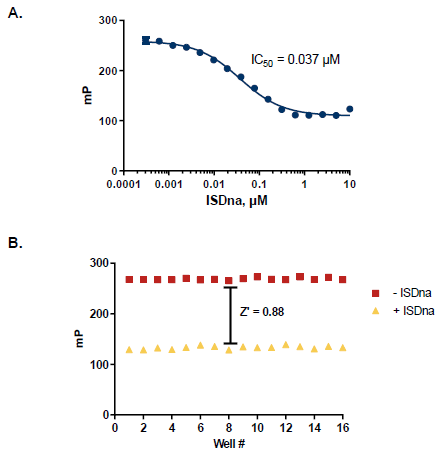
Figure 5. Assay Robustness for HTS Screening. A. ISDna dependence TREX1 reactions; half-maximal response of 37 nM. B. Z’ measurement using optimized TREX1 assay conditions (n=16). A Z’ of 0.88 demonstrates a robust assay method amenable to HTS. Image Credit: BellBrook Labs
Pilot screen of 3056 small molecules
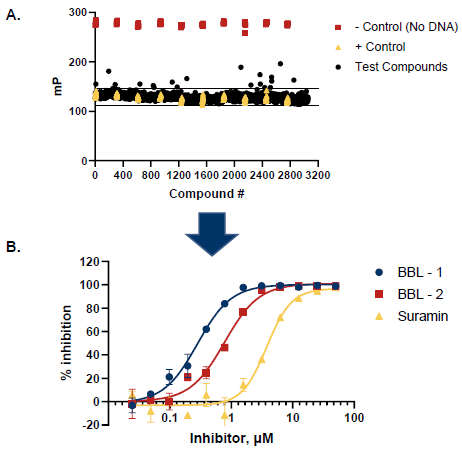
Figure 6. Pilot Screen of 3056 Small Molecules. A. 3056 compounds were screened. A total of 15 potential inhibitors were identified with activity levels 3 standard deviations above the mean. B. Selected hits from the pilot screen were tested in dose-response mode (12 concentrations, n = 2). Additionally, a dose response curve for the control Suramin is shown (IC50 = 3.0 µM). Image Credit: BellBrook Labs
Triaging and selectivity profiling
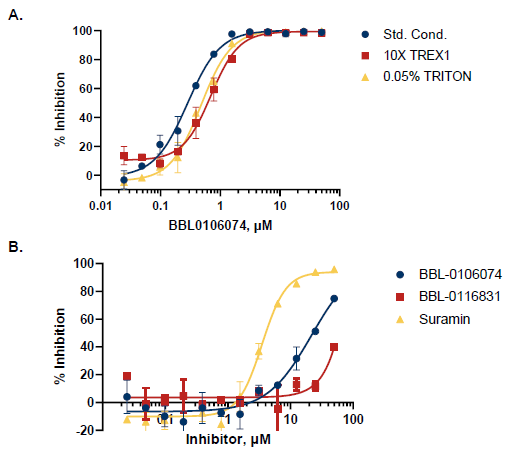
Figure 7. Triaging Assays and Selectivity Profiling. A. A selected hit was triaged for non-stoichiometric inhibitors by disaggregation assay (0.05% Triton X-100), and inhibitor titration with 10X enzyme. B. The Transcreener dAMP Exonuclease Assay allows selectivity profiling against TREX2. Both compounds are more than 50-fold less potent for TREX2, while Suramin, a positive control, inhibits TREX1 and TREX2 at similar IC50 (3.0 µM for TREX1 and 3.5 µM for TREX2). Image Credit: BellBrook Labs
Conclusions
- The Transcreener dAMP Assay is a far-red, FP homogenous immunoassay that can detect dAMP at concentrations ranging from 0.1 to 100 µM.
- The assay delivers high data quality (Z’ > 0.7) and signal (>100 mP polarization shift), allowing the detection of dAMP at nanomolar concentrations.
- Real-time readouts can be used to evaluate enzyme kinetics or inhibitor residence time.
- Pilot screens validated the assay for discovering TREX1 inhibitors, triaging hits, and profiling selectivity against comparable targets.
- The Transcreener dAMP Exonuclease Assay will assist in the quick discovery of TREX1 modulators for programs targeting TREX1 enzymes and related targets, including those downstream of the cGAS/STING Pathway
About BellBrook Labs
BellBrook Labs is dedicated to providing scientists with enabling screening tools to accelerate the discovery of more effective therapies. Leveraging its two base platforms, BellBrook has developed easy-to-use assays for hundreds of drug targets.
Transcreener® Biochemical Assay Technology
The proprietary Transcreener HTS Platform uses a highly specific antibody and far-red tracer for fluorescent immunodetection of nucleotides, including ADP, UDP, GDP, AMP, and GMP. Because its based on detection of nucleotides, the assay is universal for use with virtually any enzyme that produces these nucleotides, such as kinases, glycosyltransferases, GTPases, helicases, ATPases, nucleotidases, exonucleases and PDEs. The assay boasts direct detection of many of these enzyme targets (no coupling enzyme needed), simplyifing the protocol and reducing compound interference.
AptaFluor® Biochemical Assay Technology
AptaFluor leverages a spit aptamer technology to directly detect SAH, the common product of Methyltransferases. As the most sensitive HTS methyltransferase activity assay available, AptaFluor dramatically reduces enzyme usage and allows the assay to be run at or below Km for SAM.
Enzolution™ Assay Systems
Enzolution Assay Systems used with Transcreener Assay technology make for a comprehensive assay solution. Enzolution includes the enzyme, substrate, assay plates and buffers required to produce the enzyme reaction. Using these together simplifies researchers' assay needs without the need to spend time and money sourcing enzymes and developing assays.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.