Sponsored Content by RefeynReviewed by Olivia FrostJul 19 2024
Mass photometry is able to offer users efficient and accurate characterization of complex samples, including those that feature membrane mimetics. This technique is ideal for streamlining membrane protein purification processes, a feature highlighted by the example application of mass photometry measurements presented here.

Image Credit: stock_wichel
This case study sees measurements undertaken at various points throughout the process of purifying SMALP-embedded AQP4 samples.
Membrane proteins fulfill a range of essential biological roles, including signal transduction, immune system recognition, and molecule/ion transport across membranes.1
It is often necessary to extract, solubilize, and stabilize membrane proteins embedded in a lipid bilayer, which often involves using membrane mimetics.2
Nanodiscs are garnering increased interest in this area due to their capacity to provide improved stability and their potential for dilution to low concentrations without losing shape or integrity.
Styrene maleic acid polymer lipid particles (SMALPs) represent especially attractive options, as these function by encapsulating pieces of the lipid bilayer. This particular extraction process affords the studied proteins a native-like, detergent-free environment.3
Mass photometry and the AQP4 membrane protein
Mass photometry is a robust analytical approach that measures the mass of molecules in a solution. This technique functions rapidly and with very little required sample while being largely compatible with membrane mimetics such as nanodiscs. These factors make mass photometry an ideal addition to membrane protein purification processes.
Aquaporin-4 (AQP4) is an integral membrane protein that functions as a bidirectional water channel. AQP4 is typically present in the central nervous system (CNS), making this a promising drug target for CNS edema.4
This article looks at the purification of a sample of M1-isoform AQP4 embedded in SMALP nanodiscs as part of a functional and structural characterization process.
Mass characterization of AQP4 purification fractions
Mass photometry was utilized to investigate the composition and quality of protein samples at various points during the purification process. This experiment aimed to identify sample components and characterize isolated nanodisc-embedded AQP4 singlets before engaging in further study.
Once the embedding process had been completed, size exclusion chromatography (SEC) was employed in the separation of nanodisc-embedded AQP4 proteins from any impurities, such as empty discs or aggregates.
Mass photometry was used to characterize any fractions expected to contain the nanodisc-protein complex to identify the fraction with the greatest abundance of nanodisc-embedded AQP4 singlets (Figure 1).
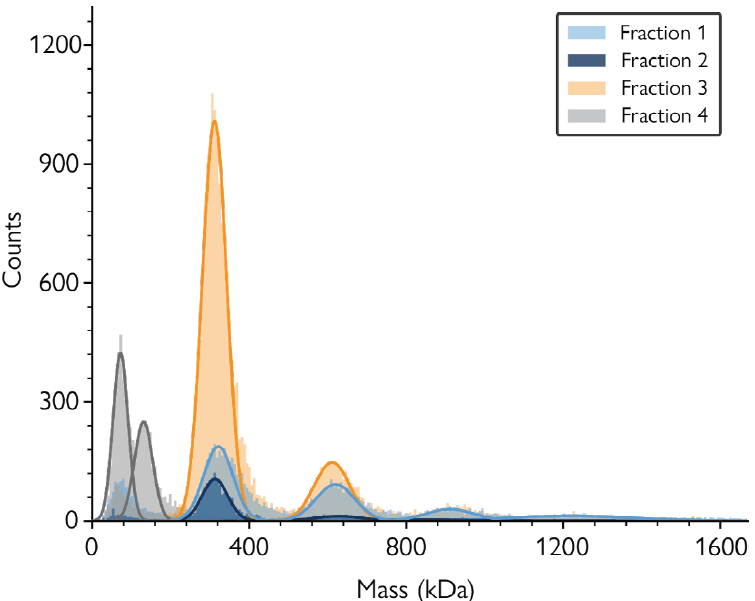
Figure 1. Mass photometry characterization of SEC fractions collected in the purification process of SMALP-embedded AQP4. Mass histograms show four selected fractions with varying proportions of the different complexes (detailed in Box 1). Fraction 3 (orange) had the highest proportion of AQP singlets (mass of ≈310 kDa). Image Credit: Refeyn Ltd.
Peak interpretation
The mass histogram peaks in Figure 1 correspond to:
≈70 and 140 kDa: Purification contaminants
≈310 kDa: One tetramer of AQP4 (singlet)
≈585 kDa: Two tetramers of AQP4 (doublet)
≈862 kDa: Three tetramers of AQP4 (triplet)
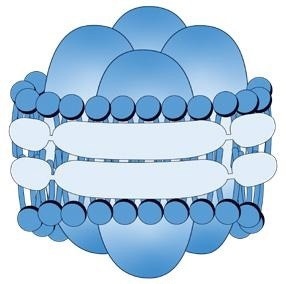
This image shows an example of a single AQP4 tetramer embedded in a SMALP (singlet). Groups of several tetramers (doublets, triplets etc.) can be found embeded in SMALPs of variable sizes. Image Credit: Refeyn Ltd.
Identification of desired species
Within this study, it was possible to confirm the presence of natively folded AQP4 singlets in the affinity-purified sample through the use of an anti-AQP4 antibody.
Analyzing the sample incubated with anti-AQP4 antibody using mass photometry illustrated the presence of a new population at ≈449 kDa, corresponding to anti-AQP4 antibodies that had bound to nanodisc-embedded AQP4 singlets (Figure 3, blue histogram).
Scattered events present in higher mass regions imply the presence of antibodies bound to doublets and triplets. Variable SMALP nanodisc masses resulted in the absence of clearly resolved peaks, however, preventing proper quantitative analysis in this instance.
A similar mass distribution to the untreated sample (Figure 1) was observed when employing negative control using a non-specific antibody (anti-GFP), though an additional peak at 145 kDa was noted, corresponding to an unbound antibody. Figure 3 shows the combination of both experiments, verifying the presence of natively-folded AQP4 in the sample.
Discussion
Mass photometry addresses many of the challenges encountered during membrane protein purification, as this analytical technique requires very small amounts of sample and is compatible with the majority of membrane mimetics.
Measurements take minutes using mass photometry, making this an ideal tool for the characterization of different fractions during purification.
SMALPs are known to have variable sizes, and this variability is reflected in the data presented here, which shows the presence of additional broad peaks in the higher mass ranges.
These peaks may represent larger complexes where relative protein and SMALP masses are unknown (Figure 2), meaning it can be especially difficult to draw conclusions about the SMALP:AQP4 proportions of higher mass components in these instances.
Masses of the complexes identified here do remain consistent with theoretical predictions, however, and the SMALP and protein masses of AQP4 singlets are in line with results acquired via other analytical methods like SEC and SMA-PAGE.5
This article showcased the ways in which mass photometry provides valuable information about complex samples, affording users greater insight into their nature and empowering them to make more informed decisions during membrane protein purification.
Experimental details
- All measurements were conducted using the TwoMP.
- Clean sample carrier slides and Refeyn 6-well sample cassettes were used.
- Samples and supplementary data were kindly provided by Philip Kitchen and Caolan Browne from Aston University.
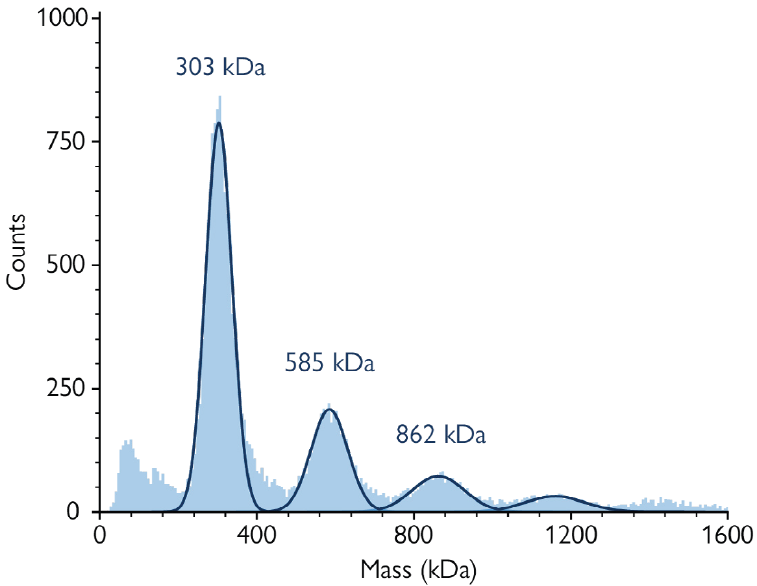
Figure 2. Affinity-purified sample of nanodisc-embedded AQP4. The mass photometry histogram shows four peaks corresponding to the main components described in Box 1. Image Credit: Refeyn Ltd.
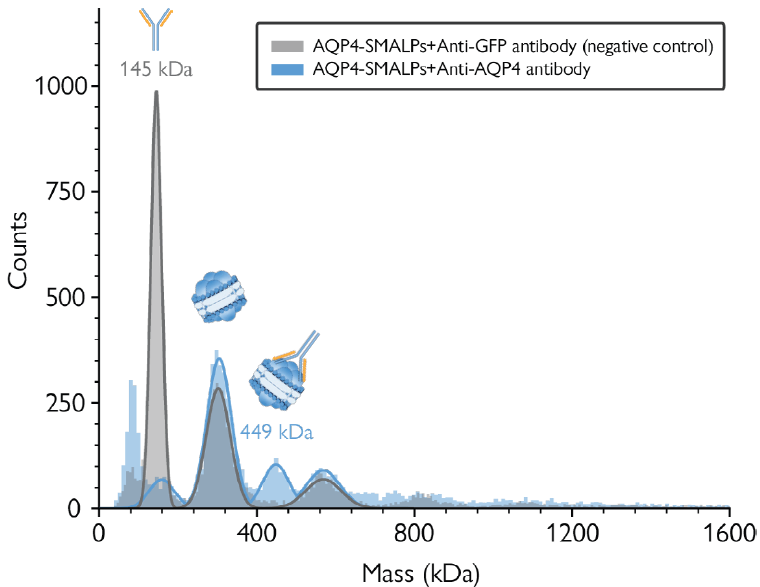
Figure 3. Antibody binding assay confirms the presence of AQP4 in the purified sample. Blue histogram: Antibody binding assay using an anti-AQP4 antibody. The peak at 449 kDa corresponds to AQP4 singlets bound to anti-AQP4 antibody. Grey histogram: Negative control with an anti-GFP antibody. The peak at 145 kDa corresponds to free anti-GFP antibody. Image Credit: Refeyn Ltd.
References and further reading
- Almeida et al., Biochim Biophys Acta Biomembr. 2017
- Majeed et al., Membranes. 2021
- Young, Biochem Soc Trans. 2021
- Kitchen et al., Cell. 2018
- Supplementary tests by Caolan Browne and Philip Kitchen
Acknowledgments
Produced from materials originally authored by Refeyn Ltd.
About Refeyn Ltd.
Refeyn are the innovators behind mass photometry, a novel biotechnology that allows users to characterise the composition, structure and dynamics of single molecules in their native environment. We are producing a disruptive generation of analytical instruments that open up new possibilities for research into biomolecular functions.
Spun out of Oxford University in 2018 by an experienced team of scientific professionals, Refeyn aims to transform bioanalytics for scientists, academic researchers, and biopharma companies around the world.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.