An interview with Shannon Cornett Ph.D., Applications Development Manager at Bruker Daltonics, discussing the use of MALDI Guided SpatialOMx for gaining deeper insight into disease research.
Why do scientists want to carry out spatial molecular analysis, particularly on diseased tissue?
We first must recognize that disease is a cellular process and in situ analysis of tissue provides the most direct and sensitive access to molecular changes occurring within cells.
The analysis of biofluids such as blood, saliva or urine has the advantage of being minimally invasive to collect, but detecting biomolecules specific to diseased cells can be complicated because of dilution and interference from products from healthy cells.
The use of spatially-oriented molecular tools such as SpatialOMx provides researchers the ability to investigate molecular changes directly at their origin in the tissue.
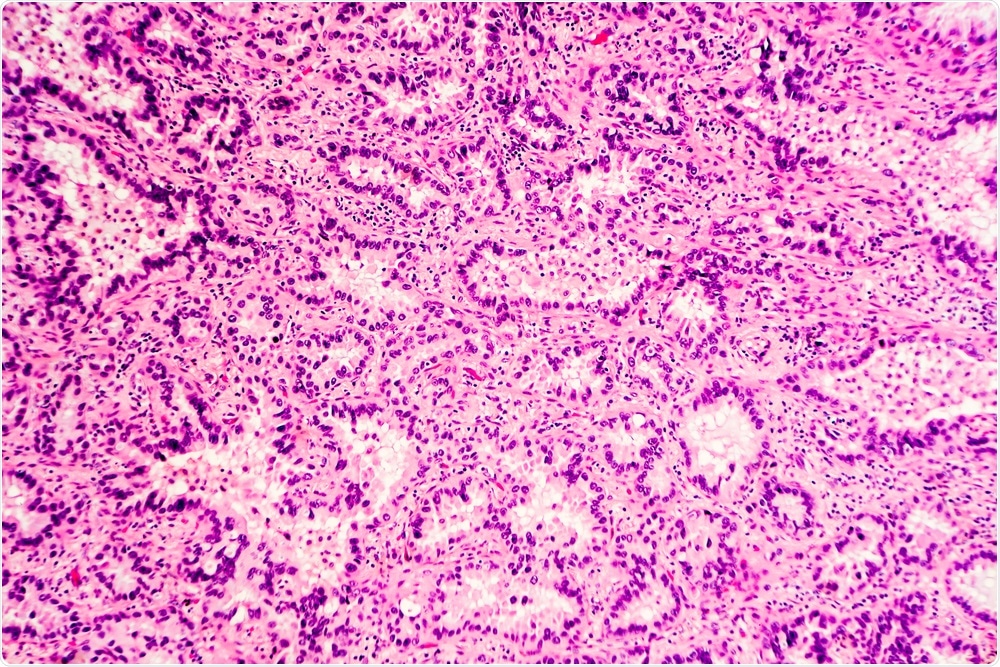 H&E stain of lung adenocarcinoma. (Image Credit: David A. Litman/Shutterstock.com)
H&E stain of lung adenocarcinoma. (Image Credit: David A. Litman/Shutterstock.com)
MALDI Guided SpatialOMx is a label-free technique that allows researchers to visualize a wide range of regionally specific molecular images in situ. Unlike antibody-based imaging techniques, SpatialOMx produces hundreds to thousands of molecular images, ranging from metabolites to proteins, for a more comprehensive investigation into disease pathways.
The current generation of commercial SpatialOMx systems approach single-cell resolution. At a routine resolution of 20 µm/pixel, the information that scientists can generate covers a wide range of compound classes and is quite specific for targeted cell clusters. This offers a much greater depth of molecular insight over existing techniques that are limited to comparatively few compounds.
How can spatially resolved data be used to better understand diseases such as cancer?
For molecular analysis, a cancer biopsy is often divided into cancerous and non-cancerous tissue. However, the cellular makeup of tissue is much more complex, being comprised of many different cell phenotypes, each carrying out unique biochemical processes. When all phenotypes are extracted into one or two solutions for molecular analysis, key molecular indicators can be lost.
SpatialOMx provides molecular fingerprints specific to cellular clusters. There have been many pilot studies published demonstrating that unique molecular fingerprints differentiate cell phenotypes, often when histology cannot. In other instances. SpatialOMx data offers very specific insight into biochemical changes occurring in regions proximal or distant to the disease.
Researchers can also use SpatialOMx to investigate how molecular histology correlates with clinical outcomes to obtain a better understanding of molecular pathways. Histologically, two biopsies may present very similar characteristics, but molecularly, there can be several different phenotypes, and this is something that has been shown in the literature.
Other clinical researchers are investigating the feasibility of constructing a classification library of molecular fingerprints versus patient history. If successful, molecular analysis may be able to help improve diagnostic and prognostic outcomes by adding a more specific molecular component to pathology.
What methods are we currently using to analyze molecular changes within tissues?
Pathologists use many techniques to analyze a tissue. One of the most common is histological staining, which has been used as a diagnostic tool for over 100 years. Here, a section of tissue is stained with a non-specific dye and a pathologist examines the stained section of tissue under the microscope and makes diagnoses based on how cell morphology compares to a grading scale. In many cases, histology does not provide a clear diagnosis because of the subjective nature of the process.
A second approach, ImmunoHistoChemical (IHC) staining, is also common. With IHC, a section of tissue is exposed to a tagged antibody specific for a known disease marker protein. The pathological diagnosis is then based on visual analysis of the antibody tag.
The nature of IHC requires knowing the specific target protein which isn’t compatible with a discovery workflow. In contrast, SpatialOMx visualizes a much broader spectrum of metabolites and lipids as well as proteins without molecular tags.
How does MALDI Guided SpatialOMx differ from these techniques, and what advantages does it offer for ‘omics analyses?
MALDI Guided SpatialOMx is enabled by Bruker's timsTOF fleX instrument. With our unique dual source, customers can now seamlessly combine results from LC-MS Omics and MALDI Imaging for unprecedented specificity and sensitivity.
MALDI Imaging guides researchers to regions of cells that present the desired molecular phenotype for performing more specific LC-MS analyses for a true SpatialOMx analysis.
If you review the scope of clinical research using mass spectrometry over the last 20 years, the 'omics (proteomics, metabolomics, lipidomics) techniques have become more and more popular and powerful.
Usually, these studies involve cohorts of clinical samples that are each extracted and analyzed by LC-MS/MS. Data is then examined to identify molecular differences across the cohort or changes that have occurred relative to the state of the original sample biopsy.
One of the advantages of traditional ‘omics analyses is that the extraction process accesses a large compliment of material from tissue specimens. However, extracting the heterogeneous tissue into a single solution blinds one to molecular contributions from any individual cell phenotype and makes it impossible to track any detected changes in molecular expression to a cellular origin.
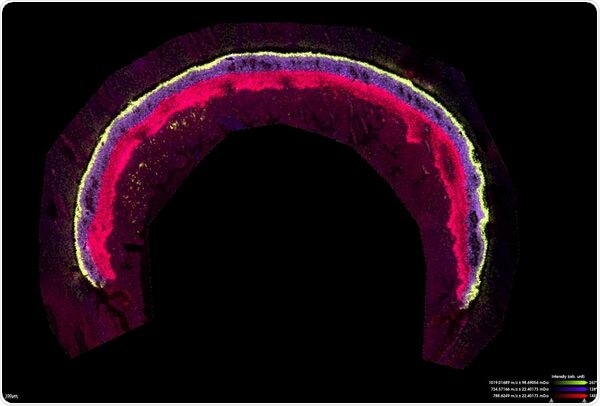 Mouse retina measured for lipids at 10 µm in zoom mode. Scale bar indicates 100 µm
Mouse retina measured for lipids at 10 µm in zoom mode. Scale bar indicates 100 µm
(Image Ctredit: Bruker Daltonics and Jeffrey Spraggins, Ph.D., Mass Spectrometry Research Center, Vanderbilt University)
Several pilot studies have combined different instruments to examine tissue cohorts using both LC-MS analyses and MALDI Imaging. With the introduction of the timsTOF fleX, our customers now have a single integrated platform for MALDI Imaging and LC-MS.
In practice, we use MALDI Imaging to identify locations of unique cell phenotypes from the MALDI Imaging data, and then target those spatial coordinates for microextraction and LC-MS/MS analysis.
Histology offers some degree of cellular selectivity, but there are numerous published examples where histology is unable to differentiate cell phenotypes, but molecular signatures derived from the MALDI Image have been shown to differentiate. MALDI Guided SpatialOMx offers the highest degree of selectivity and specificity for identifying specific cellular phenotypes and studying molecular differences between them.
The technique is also being used in the pharmaceutical industry. How does this compare to previous techniques?
Within the pharma industry, there are two obvious applications for SpatialOMx. The first is to measure the abundance and distribution of dosed therapeutic compounds in test subjects, in preclinical trials.
After dosing an animal, SpatialOMx can help determine precisely where the drug compound distributes and whether it targets the proper organ, or the diseased cells? The technique also provides a direct way of monitoring drug metabolite distributions.
Targeted SpatialOMx is possible when we know the compound(s) we are looking for. If we can detect the compound, images reveal where it is most abundant within the organ or the animal itself, and these distributions can be correlated with information from other imaging modalities. Often, metabolites of the test compound are also imaged in the same measurement.
Compared with traditional whole-body autoradiography targeted SpatialOMx offers unique advantages. In whole-body autoradiography, test animals are dosed with radio-labeled drug compounds and subsequently analyzed to determine sites of radioactivity. Usually, there is insufficient molecular specificity to determine whether the detected radioisotope is attached to an intact drug molecule or drug metabolite.
Secondarily, the cost and time for generating and handling radioactive compounds can be quite high confining these measurements to later in the drug discovery pipeline. In contrast, a targeted SpatialOMx measurement requires only a few hours and has a very low operational cost and provides a much deeper information content.
Quantifying the amount of compound present in each region of tissue is also important. LC-MS is a standard tool for quantitation from extracted tissue. The measurement yields a single amount of compound present and that is assumed to be uniformly distributed over the entire piece of tissue extracted. It is not possible to determine anything about distributions within sub-compartments of that original tissue. With targeted SpatialOMx, compound distribution within each pixel can be quantified.
A secondary benefit of SpatialOMx analysis is that the measurement that produces maps of targeted compounds and metabolites also contains distribution maps of many other compounds endogenous to the tissue, for untargeted or discovery analysis of pharmacodynamics of the drug.
Prof. Dr. Carsten Hopf talking about the timsTOF fleX
How does the timsTOF fleX differ from the timsTOF Pro? For laboratories that are currently using a timsTOF Pro, should they consider upgrading to the newer model?
As mentioned earlier, the timsTOF fleX is a timsTOF Pro with a high-speed MALDI source. So, in basic terms, the timsTOF Pro is capable of only LC-MS whereas the timsTOF fleX is capable of both LC-MS and MALDI Imaging. Further, it is important to note that timsTOF fleX requires no mechanical or configurational changes to switch between LC-MS or MALDI analyses.
For a lab that might already have a timsTOF Pro, it was most likely purchased to carry out proteomics or metabolomics. Sometimes, measurements will be done from extracts of tissue specimens, and other times, they may be using original fluid specimens.
For those researchers who plan to analyze tissue biopsies, it is important to understand the limits of common techniques, whether they involve tissue homogenizing and extraction of pieces of tissue or done following histology-guided microextraction. As already discussed, these approaches either lose the spatial information completely or lack the specificity for targeting specific molecular phenotypes of cells.
TimsTOF fleX, with the combined LC-MS and MALDI capability, enables MALDI Guided SpatialOMx which provides greater cellular specificity for performing the LC-MS extraction and analysis.
timsTOF fleX - MALDI Guided SpatialOMx on the timsTOF fleX
How do you think the field of proteomics will evolve as technologies such as the timsTOF fleX become more advanced and more widely available to researchers?
The most significant area of change to expect will be the ability to incorporate the spatial component fully into the molecular information pipeline using MALDI-guided SpatialOMx. Publications have shown that molecular phenotype can be much more specific and selective than histology alone and using SpatialOMx allows researchers to fully capitalize on the higher specificity.
At Bruker, we provide our customers with faster technologies that offer higher sensitivity and better spatial resolution. Integrate technology such as the timsTOF fleX is positioned to greatly enhance traditional workflows because if you are already doing proteomics or metabolomics to determine molecular changes, then it is a natural add-on to have that MALDI-guided capability to understand where those molecular changes are occurring. By delivering new innovative solutions, we enable researchers to take alternative, more rewarding routes.
Where can our readers find more information?
MALDI Guided SpatialOMx on the timsTOF fleX
About Shannon Cornett
 Dale Shannon Cornett, Ph.D., University of Georgia, 1993, joined Bruker in 1994 as Applications Scientist and by 2002 was Product Manager of benchtop MALDI systems. He then moved to Vanderbilt University to become a Research Assistant Professor of Biochemistry working with Professor Richard Caprioli to develop new tools and methodologies in the then-emerging field of imaging mass spectrometry.
Dale Shannon Cornett, Ph.D., University of Georgia, 1993, joined Bruker in 1994 as Applications Scientist and by 2002 was Product Manager of benchtop MALDI systems. He then moved to Vanderbilt University to become a Research Assistant Professor of Biochemistry working with Professor Richard Caprioli to develop new tools and methodologies in the then-emerging field of imaging mass spectrometry.
Shannon rejoined Bruker Daltonics in 2009 to support imaging products and now manages the MS imaging business in the Americas and Asia-Pacific. He continues to hold an appointment as Adjunct Research Professor in Biochemistry at Vanderbilt.
Please note: The products discussed here are for research use only. They are not approved for use in clinical diagnostic procedures.