Australian researchers from the Commonwealth Scientific and Industrial Research Organization (CSIRO) have found a new way to decode the genome of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes COVID-19 disease. This approach could help in drug and vaccine development. Their work titled, "Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS-CoV-2 outbreak," is published in the latest issue of the journal Transboundary and Emerging Diseases.
The researchers explain that the COVID-19 pandemic is far from over, and there is worldwide research on the development of effective diagnostic methods as well as treatments and preventable vaccines. For these research purposes, the best possible animal models need to be determined. Unless the viral genome is clearly understood and appropriate cell lines and animal models are chosen, research successes in animals could fail to be replicated in humans, the team explains.
According to CSIRO Chief Executive Dr. Larry Marshall, "The more we know about this virus, the better armed we'll be to fight it. This highly complex analysis of the genome sequence of the SARS-CoV-2 virus has already helped to determine which strains of the virus are suitable for testing vaccines underway at the Australian Centre for Disease Preparedness in Geelong - the only high biocontainment facility of its kind in the Southern Hemisphere."
What was done?
They write that this study was undertaken on the "currently chosen SARS‐CoV‐2 strains for international coronavirus disease (COVID‐19) models in the context of their phylogeny as well as in a novel alignment‐free bioinformatics approach".
For this study, they looked at published data from around the world where genome sequences have been published. This helped them fast-track their research to understand the novel coronavirus better. The researchers developed a unique visualization platform using algorithms and bioinformatics. These algorithms have been used earlier for analyzing the human genome, wrote the researchers. They used thousands of genetic sequences of SARS-CoV-2 that are now available.
CSIRO's Bioinformatics Team Leader and first author of the paper, Dr. Denis Bauer, also explained that this virus would evolve, and thus this analysis would be valuable. This genomic code lays the blueprint of the virus and its features, they wrote. Dr. Bauer, Honorary Associate Professor at Macquarie University, Australia, added, "Globally there is now a huge amount of individual virus sequences. Assessing the evolutionary distance between these data points and visualizing it helps researchers find out about the different strains of the virus - including where they came from and how they continue to evolve."
CSIRO researchers are studying the SARS-CoV-2 virus, which has a single-stranded RNA genome.
What was found?
They explained that phylogenetic trees usually reveal individual shared mutations. Using this new approach, the team made use of "genome‐wide co‐developing functionalities," developing a more flexible view of the different variations of the RNA viruses over time and sequential multiplications.
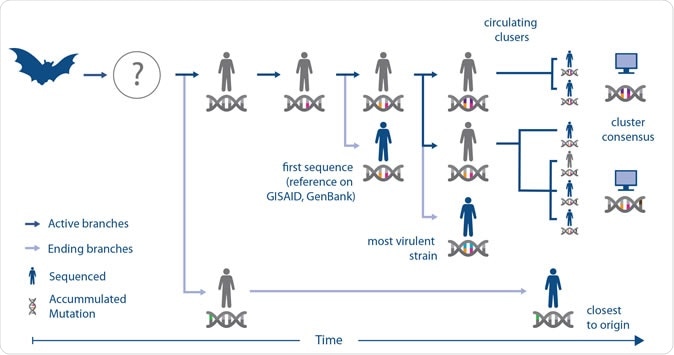
Understanding genome sequences helps researchers choose the right strain of the virus for vaccine and diagnostic efforts.
Conclusions and implications
The researchers wrote in the conclusion that the presently studied models cover only the existing virus strains. With evolutional changes, the virus may undergo mutations and variations which may not be adequately studied using these existing models.
They said, "Based on insights from the non‐discrete alignment‐free approach and experimental observations, we suggest isolates for future animal models."
CSIRO's Dangerous Pathogens Team Leader Professor S.S. Vasan, the corresponding author of this study, said that the initial 181 genomic sequences of the virus were analyzed in order to understand its behavior and impact. Professor Vasan added, "This RNA virus is expected to evolve into a number of distinct clusters that share mutations, which is what we have confirmed and visualized. At this time, we do not expect it will affect the development and evaluation of COVID-19 vaccines, therapies, and diagnostics, but it is important information to monitor as preclinical and clinical studies progress." He said, "To enable this, we are calling on the international research community to share de-identified details of case severity and outcome, and other relevant meta-data such as co-morbidities and smoking status, alongside the genomic sequences of the virus."
CSIRO's Australian e-Health Research Centre CEO David Hansen, lauded this effort saying, "Following the scientific process of peer-reviewed open publication such as this one is a vitally critical component of the CSIRO's response."
Dr. Bauer concluded, "The advantage of the data visualization platform is that it highlights evolving genetic mutations of the virus as it continues to change and adapt to new environments. The more informed we are about the genetic differences and their likely consequences on the progression of the disease, the better we can tackle the disease with diagnostics and treatments."
Journal reference:
Bauer, D.C., Tay, A.P., Wilson, L.O., Reti, D., Hosking, C., McAuley, A.J., Pharo, E., Todd, S., Stevens, V., Neave, M.J., Tachedjian, M., Drew, T.W. and Vasan, S. (2020), Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS‐CoV‐2 outbreak. Transbound Emerg Dis. Accepted Author Manuscript. doi:10.1111/tbed.13588, https://onlinelibrary.wiley.com/doi/abs/10.1111/tbed.13588