The coronavirus disease (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to spread globally. As of July 1, 2021, over 182 million cases of COVID-19 have been reported worldwide, which has led to the deaths of almost 4 million.
 Study: SARS-CoV-2 Variants are Selecting for Spike Protein Mutations that Increase Protein Stability. Image Credit: Design_Cells / Shutterstock.com
Study: SARS-CoV-2 Variants are Selecting for Spike Protein Mutations that Increase Protein Stability. Image Credit: Design_Cells / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A team of researchers at the Department of Medical Physics and Biomedical Engineering at the University College London demonstrated that mutations in the S protein of SARS-CoV-2 select for amino acid changes that result in a more stable protein than would otherwise be expected.
Based on calculating all possible protein mutations in the S protein of SARS-CoV-2, the researchers show that vast numbers of variants are more stable than expected compared to the background. The study, posted to the bioRxiv* pre-print server, highlights the importance of protein stability in understanding SARS-CoV-2 evolution.
SARS-CoV-2 mutations
Currently, new variants of concern (VOCs) threaten vaccine effectiveness, as they may evade the body's immune response. Also, these variants are easily transmitted from one person to another. Previous studies have also shown that these variants might increase the risk of severe illness and death among vulnerable populations.
Many of the SARS-CoV-2 VOCs contain mutations in the S protein that confer an advantage to the virus. The virus may have increased angiotensin-converting enzyme 2 (ACE-2) receptor binding, cleavage site alterations, and immune system evasion. The mutations may also have enhanced protein stability.
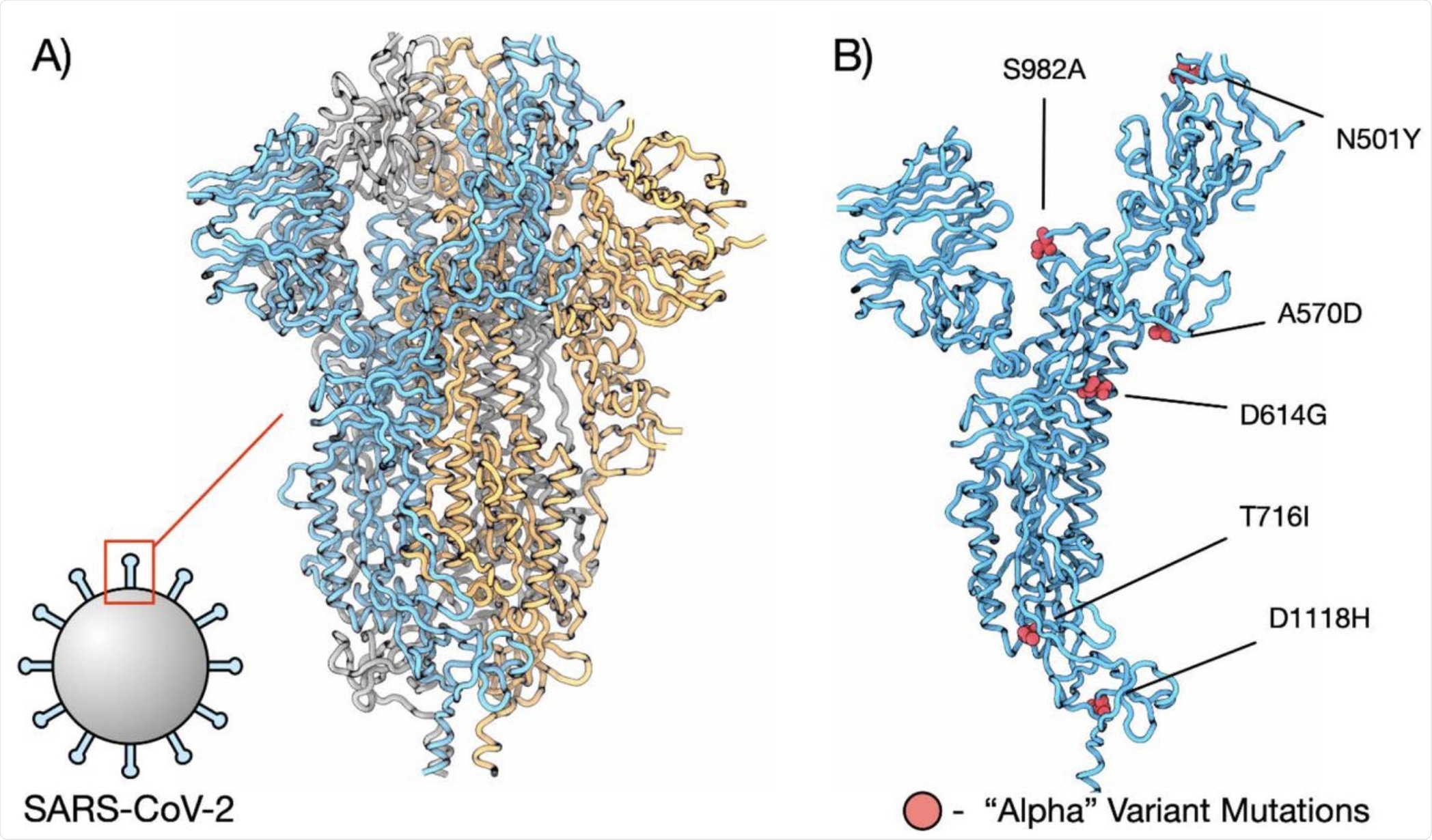 Structure of SARS-CoV-2 spike protein. A) Full structure of COVID spike protein structure (PDBID 6XVV), subunits are coloured blue, orange, and grey. B) Single subunit of spike protein with residues from the "Alpha" variant coloured red and labelled.
Structure of SARS-CoV-2 spike protein. A) Full structure of COVID spike protein structure (PDBID 6XVV), subunits are coloured blue, orange, and grey. B) Single subunit of spike protein with residues from the "Alpha" variant coloured red and labelled.
Shedding light on these properties can help determine the differences of mutations, including increased infectivity or vaccine resistance. To this end, the researchers of the current study utilized protein folding calculations as a tool to detect potentially advantageous mutations of SARS-CoV-2.
An overview of Gibbs Free Energy
Protein folding is governed by the thermodynamic principle of Gibbs Free Energy (ΔΔG), which states that the amino acid sequence of a protein determines its native structure. This native structure has the minimum ΔΔG.
When a mutation in the amino acid sequence of a protein occurs, changes in ΔΔG will also follow. In protein engineering and design, the prediction of changes in ΔΔG is routinely used for enzyme optimization or stability of protein complexes. The researchers of the current study recently demonstrated that this could function as a predictor of mutations that cause destabilization or damage to a protein in a cancer context.
ΔΔG in SARS-CoV-2 mutations determines protein stability
While the stability of mutations in the S protein of SARS-CoV-2 has been assessed in previous studies, variant analyses have not yet been conducted. SARS-CoV-2 S protein mutations are likely to improve binding to other molecules, as well as provide a longer lifespan before thermal unfolding.
Protein structural information, which has been published recently for the S protein of the SARS-CoV-2, is necessary for calculating predictions of ΔΔG values.
In their work, the researchers calculate the ΔΔG for every possible missense mutation in the SARS-CoV-2 S protein, which amounted to a total of 19,440 mutations. This information was subsequently described as the "background" mutation rate, which shows that emerging SARS-CoV-2 variants exhibit lower ΔΔG and a higher proportion of stabilizing mutations than expected.
This finding suggests that protein stability, while rare, plays a vital role in the evolution of SARS-CoV-2. Furthermore, the combination of mutations may result in synergistic changes to increase stability, revealing possible evolutionary orders for mutation.
How ΔΔG relates to SARS-CoV-2 VOCs
Taking their research one step further, the researchers compared the background mutational distribution between VOCs and SARS-CoV-2 “Variants of Interest (VOIs).” Both SARS-CoV-2 VOIs and VOCs were found to exhibit significantly lower ΔΔG values as compared to the bulk population, thereby indicating that these variants likely have been evolutionarily selected for stabilizing or non-destabilizing mutations.
Notably, none of the VOCs were found to contain mutations that were strongly destabilizing, which was defined as ΔΔG values greater than 2.5 Kcal/mol. Overall, the researchers concluded that a statistical enrichment exists for SARS-CoV-2 mutations that stabilize the S protein as compared to the background.
Conclusion
"This work highlights those mutations with a stabilizing effect on the SARS-CoV-2 spike protein are one of the key drivers of the evolution of the virus and contributing to the increased transmissibility of emerging variants."
Further investigation is needed to study the stability of the SARS-CoV-2 S protein. The team suggests that folding simulations of newly sequenced variants may provide an affordable and modern method to highlight advantageous mutations. Additionally, a prospective simulation of further mutation may help predict future variants to emerge.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references: