In order to combat the effect of any novel virus, it is important to understand the mechanism by which it infects host cells, overcomes the host immune defense mechanism, and enhances the synthesis and replication of viral proteins.
The most prominent preventive approach to minimize the effect of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in host cells has thus far been the development of vaccines. These vaccines target the viral spike protein responsible for binding to the host cells and entering the systemic circulation.
However, vaccination does not completely stop disease propagation. As SARS-CoV-2 is a novel virus, it is inevitable that it will undergo multiple rounds of mutations that produce new variants that may be resistant to existing vaccines.
This highlights the need for the development of drugs that target the core machinery of SARS-CoV-2 and can be used to treat infected individuals. To this end, the non-structural protein 1 (NSP1) appears to be an ideal candidate as a target. Previous research has shown NSP1 to be a conserved region in all beta-coronaviruses, the family to which SARS-CoV-2 belongs.
However, previous reports in the literature about the methods by which NSP-1 invades the host system stops the synthesis of host-specific proteins and induces the synthesis of viral proteins remains contradictory. In a recent study published on the preprint server, bioRxiv*, researchers try to resolve this confusion by studying the molecular aspects of host cell entry and inhibition of host-specific cellular mechanisms.
What is NSP1?
NSP1 is an important virulence factor that plays a crucial role in the pathogenicity of SARS-CoV-2 by helping the virus evade the host innate immune response. NSP1 both downregulates the expression of host genes and promotes its own propagation by neutralizing the antiviral responses of host cells to which the virus binds.
The genome of SARS-CoV-2 is an approxiamtely 30 kilobase (kb) positive-stranded ribonucleic acid (RNA) virus with 5′-cap structure, 5′UTR (or leader), 3′UTR, and polyA tail. SARS-CoV-2 contains 10 protein-coding open reading frames (ORFs), which are parts of the genome where protein translation can occur without encountering any stop codons.
Upon cell entry, the genomic RNA (gRNA) is translated into polyprotein, which is processed into 16 non-structural proteins (NSPs). Subsequently, its gRNA serves as a template to generate a set of subgenomic mRNAs (sgRNAs) that encode for other viral proteins.
The first protein produced by coronaviruses upon infection is NSP1, which is encoded by ORF1a at the 5′ end of gRNA. In earlier studies on the related SARS-CoV-1, NSP1 has been found to inhibit host immune responses by repressing expression of host transcripts and preventing full induction of interferon (IFN) and STAT1 phosphorylation, both of which are crucial for the initiatiation of antiviral immune responses.
About the study
In the current study, the researchers assessed the potential of NSP1 as a target by studying the detailed molecular mechanisms of its functions. To this end, they synthesized the viral genome by cloning known sequences from the SARS-CoV-1 and 2 and separating specific proteins using Western Blotting techniques.
The researchers used mutagenesis to detect the effect of different mutations in specific genes to assess their roles in immune evasion by SARS-CoV-2 NSP1. Reporter assays were also utilized to detect specific changes as a result of the induced mutations. Mass spectroscopy was used to analyze the proteins.
Specific biotin-based assays (BioID) were used to locate proteins of interest and study the different proteins in the functional parts of the SARS-CoV-2 NSP1 protein. This assay helped the researchers understand the different parts of the NSP1 protein engaged in different antiviral defense mechanisms. For all these experiments, the researchers used Human HEK293T cells as hosts.
Study results
The researchers of the current study found that stem loop-1 (SL1), which is a hairpin structure in single-stranded RNA that helps the viral mRNA retain its structure and continue specific enzymatic interactions, is both necessary and sufficient to escape NSP1-mediated repression. Mutagenesis was used to isolate 3 specific cytosine proteins, which is one of the four nucleic acid bases of RNA, that were crucial to evasion of host defense mechanisms.
Upon further analysis of the protein by BioID, NSP1 was found to block host translation by inserting its C-terminal domain into the mRNA entry tunnel on the ribosomal 40S subunit. In doing so, NSP1 blocked ribosomal host-protein synthesis and exported the viral mRNAs into the system. An artificially constructed non-functional mutant KH164AANSP1 could not replicate the mechanism of the wild-type NSP1 as effectively.
Normally, when a virus attacks the host cells, viral RNA is sensed by host RNA helicases in infected cells, resulting in the activation of the transcription factors ATF2/c-Jun, IRF3/IRF7, and NF-kB. These factors subsequently induce the production of cytokines, including members of the IFN family, which go on bind to their respective receptors and trigger a second wave of cytokine signaling. These two waves upregulate genes that inhibit viral replication.
Ultimately, NSP-1 was found to evade these pathways to prevent the induction of cytokines, thereby helping the virus to spread in the host system.
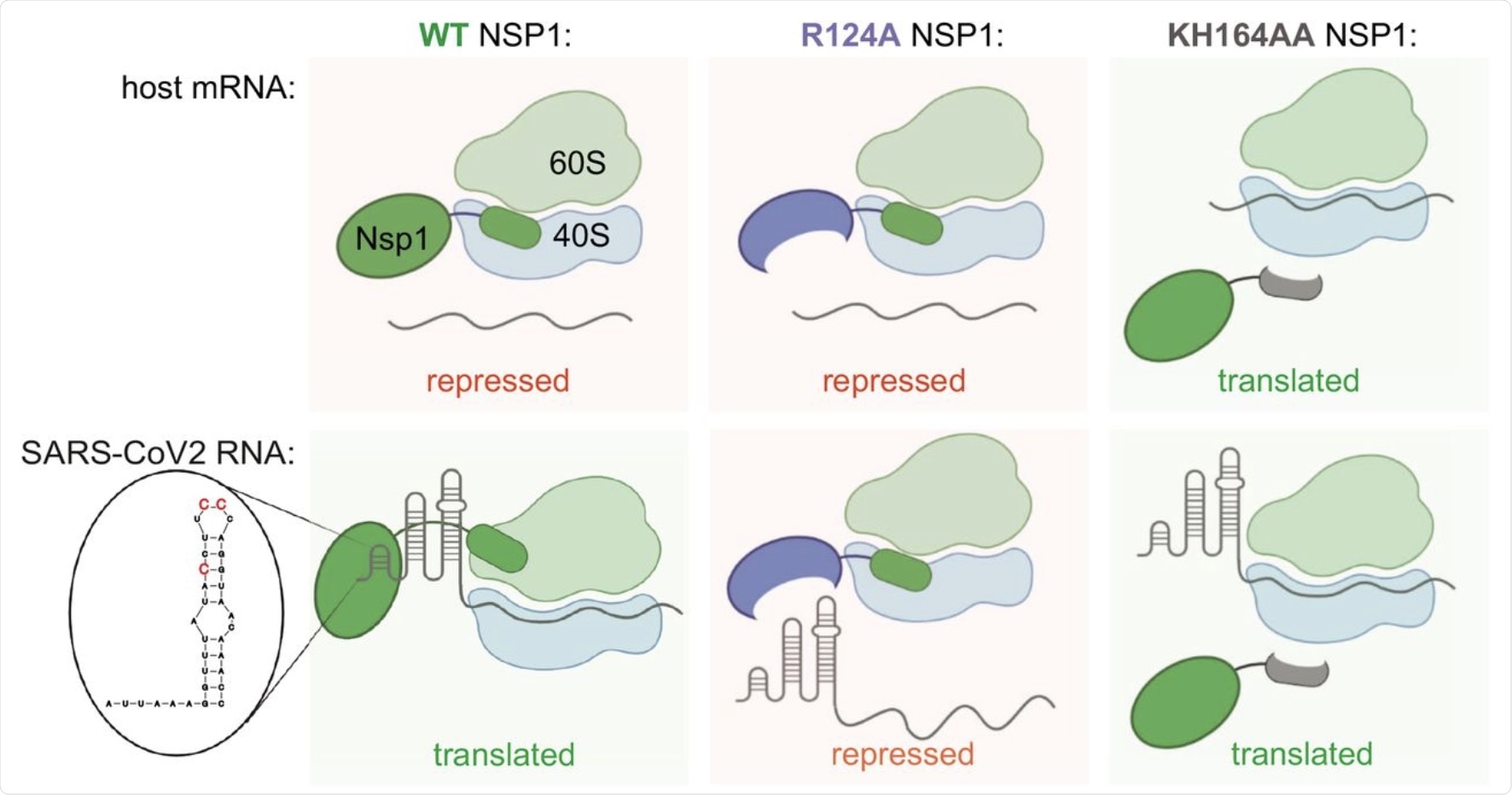 Speculative model for NSP1 function in repression of host mRNA and activation of SARS-CoV-2 expression. WT NSP1 protein represses translation of host mRNA via blocking the ribosome entry tunnel. SARS-CoV-2 genomic and subgenomic RNAs escape repression by NSP1 due to SL1 in their 5′UTR, interacting with NSP1. This way viral RNA highjacks host ribosomes without competing for limiting eIFs in infected cells. In particular, positions C15, C19 and C20 are required for alleviation of NSP1 silencing. Positions K164H165 in NSP1 are required for interaction with ribosome, therefore KH164AA mutant of NSP1 does not repress either host or SARS-CoV-2 mRNA. Position R124 is required for interaction with SL1, therefore NSP1 R124A mutant represses both host and viral mRNA.
Speculative model for NSP1 function in repression of host mRNA and activation of SARS-CoV-2 expression. WT NSP1 protein represses translation of host mRNA via blocking the ribosome entry tunnel. SARS-CoV-2 genomic and subgenomic RNAs escape repression by NSP1 due to SL1 in their 5′UTR, interacting with NSP1. This way viral RNA highjacks host ribosomes without competing for limiting eIFs in infected cells. In particular, positions C15, C19 and C20 are required for alleviation of NSP1 silencing. Positions K164H165 in NSP1 are required for interaction with ribosome, therefore KH164AA mutant of NSP1 does not repress either host or SARS-CoV-2 mRNA. Position R124 is required for interaction with SL1, therefore NSP1 R124A mutant represses both host and viral mRNA.
Implications
The current study showed that the wild-type SARS-CoV NSP1 was responsible for binding to host ribosomes and inducing the production of viral mRNA. NSP1 was also found to evade immune responses and defer the release of inflammatory cytokines.
The first therapeutic target identified in this study is the binding site for NSP1 to the host ribosome, . A second target would involve inducing mutations in the NSP1 region that interacts with the viral leader to evade host antiviral responses, and that in the SL-1 region, targeting the 3 specific cytosine molecules.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bujanic, L., Shevchuk, O., von Kügelgen, N., et al. (2021). The key features of SARS-CoV-2 leader and NSP1 required for viral escape of NSP1-mediated repression. bioRxiv. doi:10.1101/2021.09.13.460054. http://biorxiv.org/lookup/doi/10.1101/2021.09.13.460054.
- Peer reviewed and published scientific report.
Bujanic, Lucija, Olga Shevchuk, Nicolai von Kügelgen, Anna Kalinina, Katarzyna Ludwik, David Koppstein, Nadja Zerna, Albert Sickmann, and Marina Chekulaeva. 2022. “The Key Features of SARS-CoV-2 Leader and NSP1 Required for Viral Escape of NSP1-Mediated Repression.” RNA 28 (5): 766–79. https://doi.org/10.1261/rna.079086.121. https://rnajournal.cshlp.org/content/28/5/766.