The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 Omicron (B.1.1.529.1) variant was first reported on November 24, 2021, in South Africa. Almost immediately, researchers recognized the danger of this variant and its potential to rapidly spread across the globe.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Even among those who have been fully vaccinated against the coronavirus disease 2019 (COVID-19), the pace with which the Omicron variant is currently spreading is of significant concern.
The Omicron variant spike protein contains three-five times the number of mutations found in prior SARS-CoV-2 strains. Understanding the implications of these mutations for angiotensin-converting enzyme 2 (ACE2) receptor binding and neutralizing antibody evasion is crucial in assisting the development of effective therapies to stop the spread of the Omicron and other future SARS-CoV-2 variants.
As compared to the initial SARS-CoV-2 wild-type strain, the Omicron variant includes 37 mutations in the spike protein, 15 of which are in the receptor-binding region (RBD). The RBD is present within the spike protein that facilitates both ACE2 receptor-mediated attachment to human cells and is the principal target of neutralizing antibodies.
The SARS-CoV-2 Delta variant, which was the dominant circulating strain of SARS-CoV-2 until the Omicron strain was detected, has seven changes in the spike protein compared to the Wuhan strain. However, only two of these mutations including the T478K and D614G are shared by the Omicron strain.
In a recent study published on the preprint server bioRxiv*, researchers from multiple Canadian institutions used surface plasmon resonance tests to compare the obtained binding affinities (KD) to wild-type and Delta spike proteins to determine the possible impact of Omicron mutations on ACE2 binding affinity of its spike protein.
In the current study, the term wild-type (WT) refers to the ancestral Wuhan-1 strain with the D614G mutation. Although the SARS-CoV-2 Omicron spike protein has a greater affinity for ACE2 than the ancestral Wuhan strain, the ACE2 affinity for Delta and Omicron variants is identical. The K417N mutation in the Omicron version is known to drastically diminish ACE2 binding.
About the study
The ACE2-Omicron spike protein complex has a high density of ACE2 attached to the RBD of one of the protomers in the "up" position according to cryo-electron microscopy (cryo-EM) structural analysis. A second bound ACE2 has a lower density, which implies that the second RBD is partially occupied under the conditions of the experiment.
The stoichiometry of ACE2 binding can vary depending on the experimental settings. In this study, the authors focused on the structure of the ACE2-spike protein interface in the ACE2 molecule that was most strongly bound. A density map with a resolution of 2.66 Å was created at the spike protein-ACE2 interface after focused refining of the RBD-ACE2 area, thereby permitting identification of sidechains participating in the interaction.
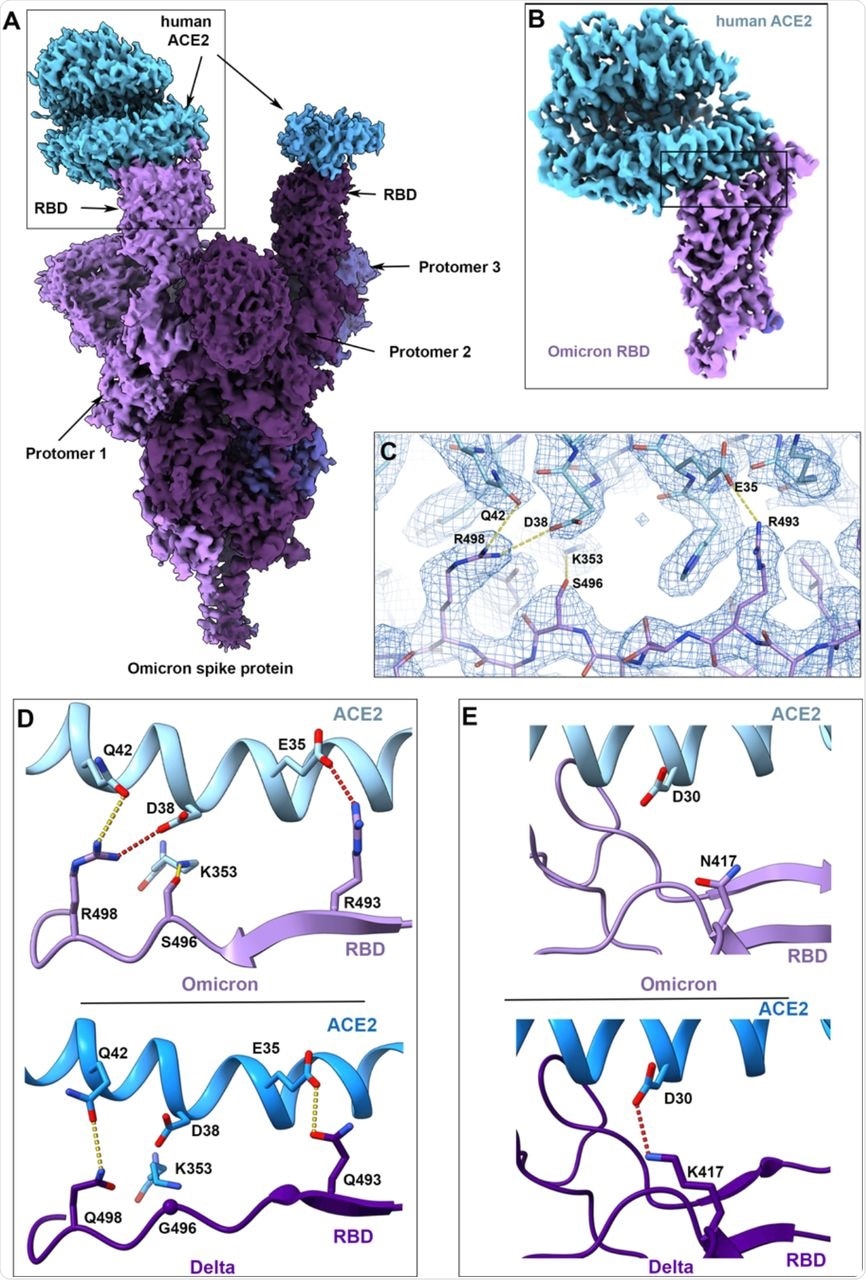
Cryo-EM structure of the Omicron spike protein-ACE2 complex.
The Q493 and Q498 residues on the spike protein create hydrogen bonds with ACE2 residues E35 and Q42, respectively, in the Delta variant-ACE2 complex. Three mutations are found in this region in the Omicron variant including Q493R, G396S, and Q498R.
The residue R493 forms a new salt bridge with ACE2 residue E35. Comparatively, residue R498 forms a new salt bridge with ACE2 residue D38 while also preserving a hydrogen-bonded connection between residue 498 and ACE2 residue Q42. By creating a hydrogen bond with ACE2 residue K353, the RBD residue S496 provides a novel interaction at the interface.
The Omicron variant of SARS-CoV-2, as compared to the Alpha, Beta, Gamma, Kappa, Epsilon, and Delta variants, showed considerable evasion from every antibody in the panel utilized in this study, with total escape from five of the six antibodies tested. Both N-terminal domain- (NTD) directed antibodies neutralizing efficacy is fully lost in the Omicron variant, as previously shown in the Alpha variant, which has identical or comparable deletions in its NTD.
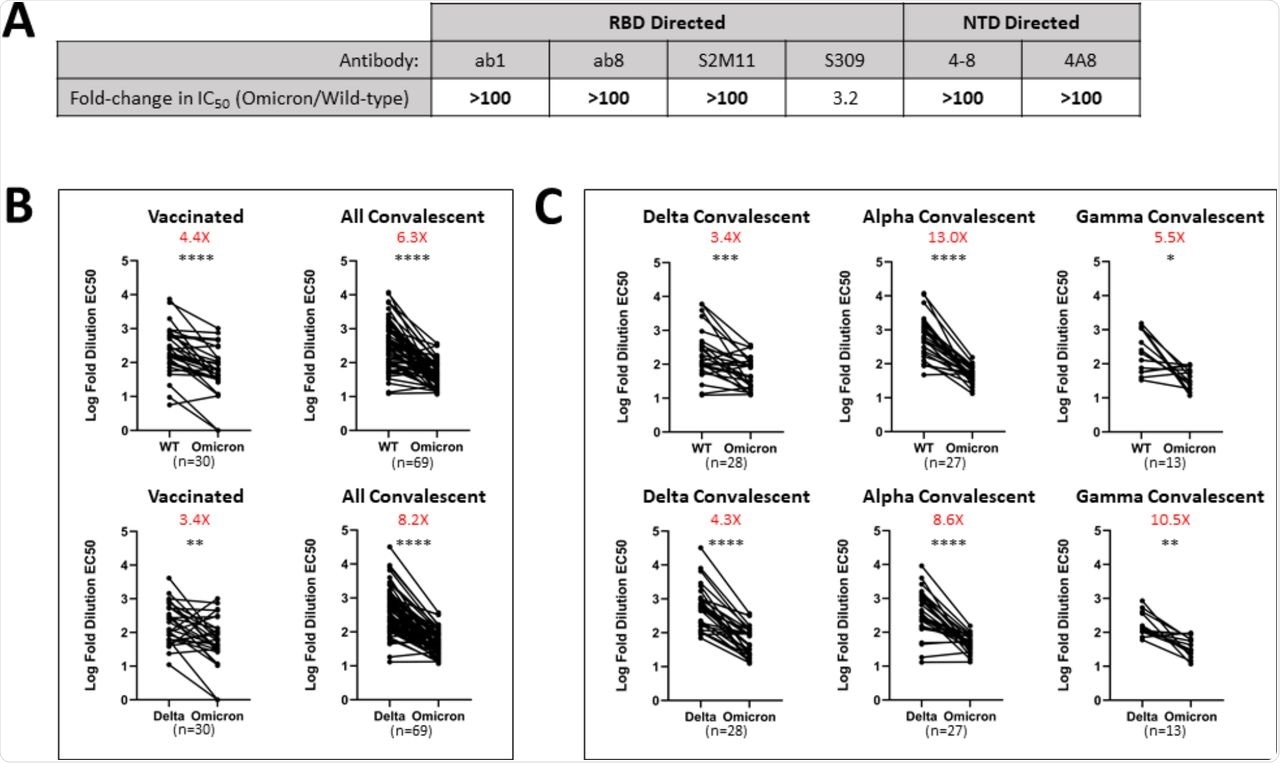
Monoclonal antibodies and vaccinated and convalescent patient-derived sera exhibit decreased Omicron neutralization potency.
Although there is still a four-fold drop in neutralization potency compared to the original strain, the Omicron variant does not show total escape from S309, which is an antibody currently being evaluated in clinical trials to treat patients with COVID-19. As compared to previously revealed variations of SARS-CoV-2, the extremely large number of mutations in the Omicron variant spike protein appear to confer unprecedently broad antibody escape.
The ability of convalescent patients' sera to neutralize the Omicron variant was found to be 6.3 times lower than the wild-type. Sera from the vaccinated cohort similarly had lower neutralization capacity, with a larger range of results-driven by those people who had a very high loss of neutralization ability to Omicron. Given Delta's global dominance, a comparable change in neutralization potential between Delta and Omicron variants is a more meaningful comparison.
Sera from convalescent patients exhibited a higher loss in neutralizing potency as compared to the Delta variant, whereas the vaccinated group has a lower reduction in potency.
Implications
Given the possibility that the newly discovered Omicron variant will quickly become the dominant circulating strain of SARS-CoV-2, the processes behind its rapid dissemination are of critical importance. The enormous number of changes on the spike protein's surface, particularly the immunodominant RBD, should help the virus evade antibodies induced by vaccination or previous infection.
However, despite these substantial alterations, the Omicron variant has evolved to retain its capacity to bind ACE2 efficiently. The cryoEM structure of the spike protein-ACE2 complex demonstrated that interactions involving the novel Omicron variant mutations at residues 493, 496, and 498 appear to restore ACE2 binding efficiency that would otherwise be lost due to mutations like K417N.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mannar, D., Saville, J. W., Zhu, X., et al. (2021). SARS-CoV-2 Omicron Variant: ACE2 Binding, Cryo-EM Structure of Spike Protein-ACE2 Complex and Antibody Evasion. bioRxiv. doi:10.1101/2021.12.19.473380. https://www.biorxiv.org/content/10.1101/2021.12.19.473380v1.
- Peer reviewed and published scientific report.
Mannar, Dhiraj, James W. Saville, Xing Zhu, Shanti S. Srivastava, Alison M. Berezuk, Katharine S. Tuttle, Ana Citlali Marquez, Inna Sekirov, and Sriram Subramaniam. 2022. “SARS-CoV-2 Omicron Variant: Antibody Evasion and Cryo-EM Structure of Spike Protein–ACE2 Complex.” Science 375 (6582). https://doi.org/10.1126/science.abn7760. https://www.science.org/doi/10.1126/science.abn7760.