There is a strong link between pathophysiology and the human body's microbiome. Research suggests that many microbial communities regulate health, host homeostasis, disease progression, and infection risk. Recent developments in high throughput sequencing technologies like metagenomic sequencing, the whole genome shotgun, and next-generation sequencing (NGS) techniques have stimulated further research on the human microbiome. This helps in the accurate detection of resident microbial DNA as well as reveals the diversity of hidden microbes on the exposed human body surfaces.
Understanding the composition of healthy male genital mucosa microbiome and its variations is essential since it might contain determinants such as risk factors for viral and bacterial infections. A few studies have assessed the male reproductive tract microbiome's role but focus mainly on the coronal sulcus or urine.
A new study in the journal Microorganisms aimed to identify and analyze studies on the composition of the microbial community on different parts of the healthy and diseased male genital mucosa.
 Study: Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Image Credit: Ebrahim Lotfi / Shutterstock
Study: Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Image Credit: Ebrahim Lotfi / Shutterstock
About the study
The study searched Google Scholar, Scopus, Web of Science, and MEDLINE (PubMed) on 2nd September 2022 for definitive keywords such as genital mucosa, male, genital tract, microbiome, coronal sulcus, and others. Clinical reports and articles published till August 2022 in English were included in the study. These studies had to include culture-independent techniques of the detected microbiota and related to the microbiome research of the genital mucosa.
Subsequently, abstracts and articles were independently selected by two authors, followed by full-text assessments. A third author resolved any disagreements concerning the eligibility of the studies. Finally, data extraction took place from the included studies to assess the quality of the studies as per the Cochrane risk-of-bias tool for randomized trials and the National Institutes of Health (NIH) Study Quality Assessment Tools for observational studies.
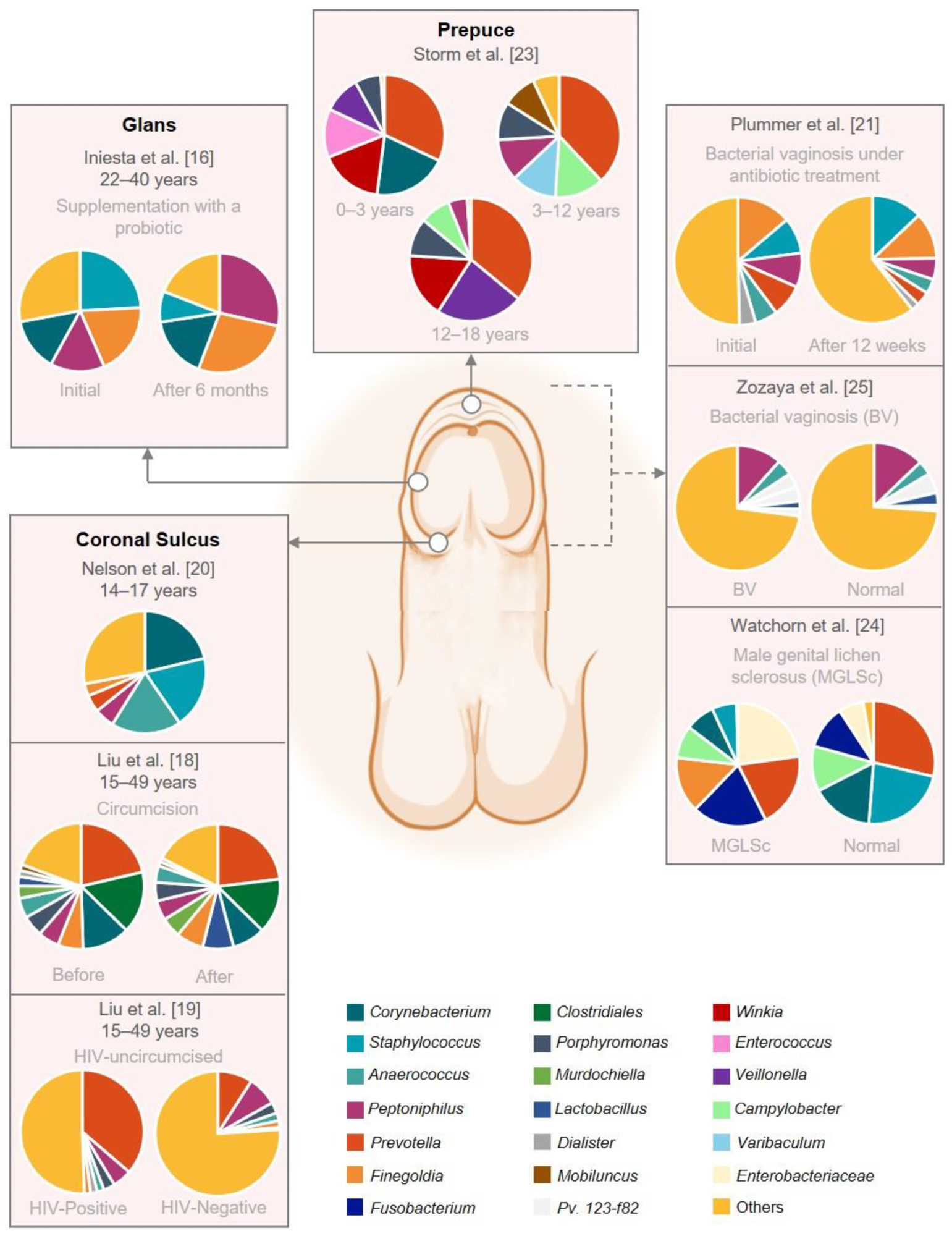 Graphical representation of the male genital mucosa microbiome composition. This figure was based on eight studies that reported the abundance on genus level. The remaining studies did not have raw abundance data available. The figure was created with BioRender.com (accessed on 16 September 2022).
Graphical representation of the male genital mucosa microbiome composition. This figure was based on eight studies that reported the abundance on genus level. The remaining studies did not have raw abundance data available. The figure was created with BioRender.com (accessed on 16 September 2022).
Study findings
The results indicated that a total of 10 studies met the inclusion criteria, and all of them were observed to employ 16S rRNA gene amplicon sequencing for male genital mucosa microbiome analysis. However, the regions of amplifications of the 16S rRNA genes differed. Analysis of the foreskin microbiome indicated no significant changes in the foreskin and urethral microbiome composition over time. Differences concerning the perineal microbiome were observed between post-pubertal and prepubertal/toilet-trained males.
A higher abundance of Winkia, Mobiluncus, Peptoniphilus, Corynebacterium, Staphylococcus, and Prevotella was reported in the foreskin microbiome. The perineum of pre-toilet-trained boys showed higher numbers of Enterococcus, Bifidobacterium, and Veillonella. Prepubertal boys showed higher numbers of Finegoldia sp., Faecalibacterium sp., Anaerococcus sp., and Peptoniphilus sp. Corynebacterium spp. was reported to be most abundant in post-pubertal boys, with S. aureus and C. tuberculosteraricum being the most common species.
The results also reported differences in urine and coronal sulcus microbiome composition, with the latter being more influenced by circumcision. The coronal sulcus comprised higher proportions of Porphyromonas, Finegoldia, Prevotella, Peptoniphilus, Anaerococcus, Staphylococcus, and Corynebacteria, while Veillonella, Gardnerella, Lactobacillus, and Streptococcus were higher in urine. Bacterial vaginosis-associated taxa were also identified in the coronal sulcus of sexually inexperienced and experienced men. Moreover, the coronal sulcus microbiome was observed to be more heterogenous before circumcision as compared to after circumcision. Also, uncircumcised men infected by HIV during a 2-year clinical trial reported higher levels of penile anaerobes than uncircumcised men who remained HIV-negative.
Furthermore, the penile skin diversity of males with women partners with bacterial vaginosis was reported to be higher, suggesting that the transmission of bacterial vaginosis-associated bacteria is common during sex. Antibiotic treatment of the male partners was observed to cure bacterial vaginosis in women. Differences in genital mucosa microbiome were observed between men with balanoposthitis and healthy men with the prepuce. Prevotella bivia and Staphylococcus warneri were reported to be the dominant species in men with balanoposthitis, while Porphyromonas somerae and Ezakiella sp. were more prevalent in men with the prepuce. Additionally, Fusobacterium, Prevotella, and Enterobacteriaceae were ported to be most abundant in men male genital lichen sclerosus (MGLSc). Finally, probiotic supplementation with L. salivarius PS116610 in couples with infertility was reported to improve the urogenital tract microbiome and solve dysbiosis in the majority of couples under assisted reproduction treatment.
Therefore, the current study demonstrates that the composition of the male genital mucosa is highly diverse. Different types of sexual activities can result in the colonization of the urethra and penis by different microorganisms. Further studies are required to understand the parasitome, virome, and mycobiome of the male genital mucosa to understand the risk factors of various infections and develop diagnosis, management, and prevention strategies.
Limitations
The current study has certain limitations. First, the sample population of most of the included studies is relatively low. Second, the data on the ethnicity of participants is most often absent. Third, most of the studies included individuals with different specialties. Fourth, the analysis across age groups of males was limited. Fifth, direct comparison between included studies could not take place. Finally, none of the included studies analyzed parasitome, virome, and mycobiome in the male genital mucosa.