A team of international scientists has recently revealed that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interacts with two heme metabolites, bilirubin and biliverdin, to escape the antibody-mediated host immune responses. The study is currently available on the medRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The cellular entry of SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), is initiated by the interaction between the viral spike protein and the host angiotensin-converting enzyme 2 (ACE2). The spike protein is the most immunogenic component of SARS-CoV-2, and thus, the most potent target of neutralizing antibodies. While it is known that the receptor-binding domain (RBD) at the C-terminal end of spike S1 subunit interacts with ACE2, the structure and function of the spike N-terminal domain is largely unknown.
In the current study, the scientists aimed to evaluate the immunogenic properties of the N-terminal domain of the SARS-CoV-2 spike protein.
Important observations
As a part of the serological analysis of SARS-CoV-2, the scientists produced a range of recombinant spike antigens using human cell lines. During the analysis of these constructs, the scientists noticed a distinctive spectrum of trimeric spike and S1 subunit that is similar to the spectrum of a heme metabolite, biliverdin. To investigate further, they isolated the pigment from the S1 subunit and confirmed the presence of biliverdin by mass spectrometry.
Biliverdin is the initial breakdown product of heme, which is further reduced to produce bilirubin. Using surface plasmon resonance, they examined the binding of bilirubin and biliverdin to the spike S1 subunit and observed that spike S1 has a strong affinity for biliverdin.
By conducting structural analysis using cryo-electron microscopy, they observed that biliverdin is positioned within a deep cleft on the spike N-terminal domain. Further analysis revealed that the binding pocket of biliverdin is surrounded by hydrophobic residues that interact with the ligand.
Because a histidine residue is present in the biliverdin binding pocket, the scientists hypothesized that the interaction between S1 and biliverdin may be pH-dependent. In line with the hypothesis, they noticed a significant reduction in biliverdin content of the S1 subunit under acidic pH. Interestingly, the substitution of amino acids closely involved in ligand binding resulted in 2- to 3-fold reduction in biliverdin-S1 binding affinity. However, they observed that the SARS-CoV-2 spike containing these amino acid substitutions is still capable of infecting cells, indicating that biliverdin binding is not associated with viral entry.
To investigate the immunological consequences of biliverdin-S1 binding, they measured the reactivity of anti-SARS-CoV-2 human sera with wild-type and N121Q mutated spike protein using flow cytometry. The findings revealed that biliverdin significantly reduces the binding of anti-SARS-CoV-2 IgG antibodies to the wild-type spike protein. However, biliverdin could not change the affinity of antibodies toward N121Q mutated spike protein.
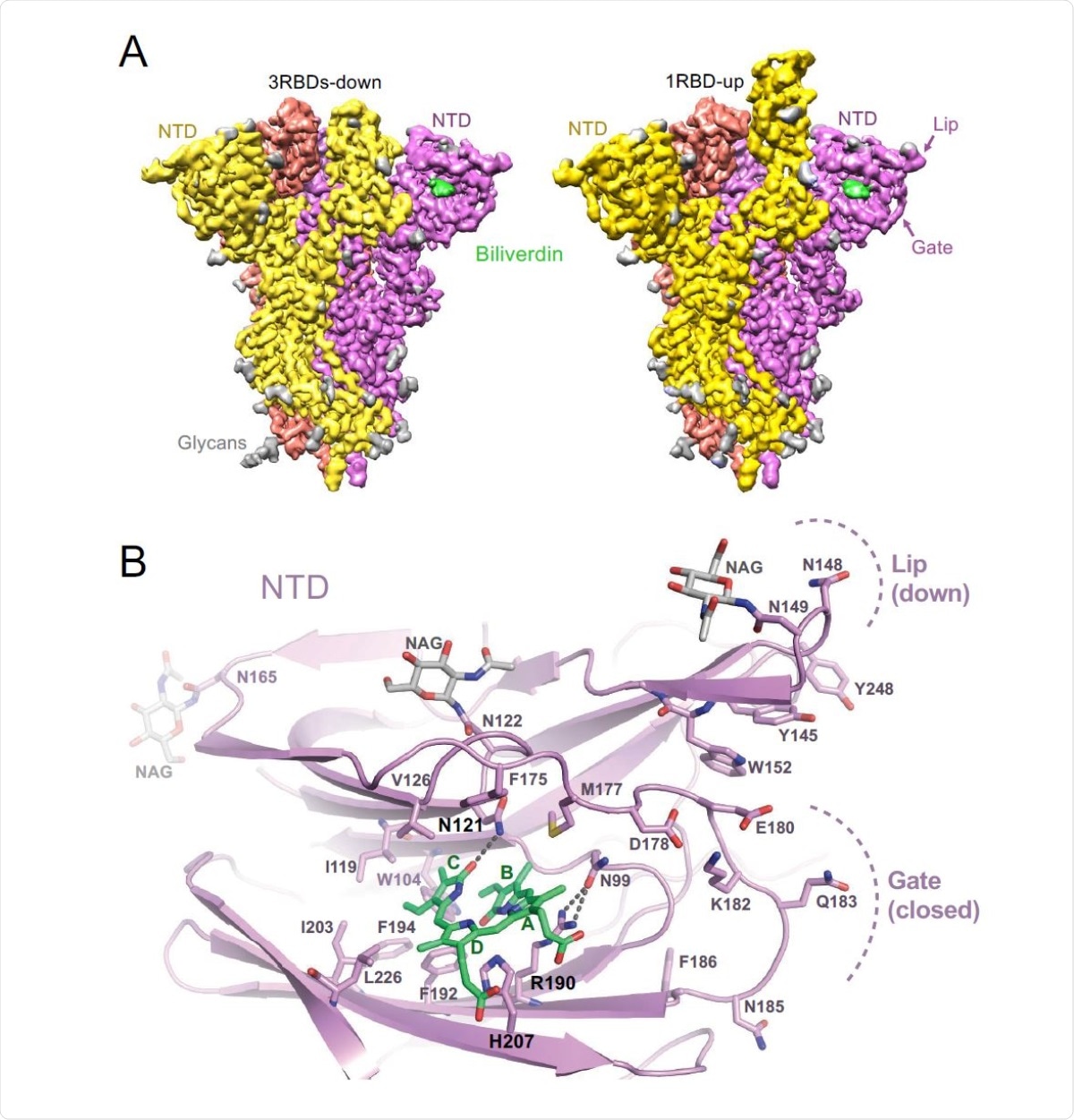
Structures of SARS-CoV-2 spike-biliverdin (A,B) and spike-P008_056 Fab (C) complexes. (A) Cryo-EM 3D reconstructions of trimeric SARS-CoV-2 spike ectodomain in 3RBD-down (left) and 1RBD-up (right) conformations determined under saturation with biliverdin. Spike protomers are color-coded. Biliverdin and glycans are shown in green and grey, respectively. (B) Details of the biliverdin binding pocket in the crystal structure. SARS-CoV-2 NTD is shown as cartoons with selected amino acid residues and biliverdin in sticks. Carbon atoms of the protein chain, sugars (NAG), and biliverdin are in purple, grey and green, respectively; the remaining atoms are coloured as follows: oxygen, red; nitrogen, blue; and sulphur, yellow. Dark grey dashes are hydrogen bonds.
Based on this remarkable observation on antigen-antibody binding, they further evaluated the binding affinity of a panel of anti-SARS-CoV-2 IgGs cloned from B cells of COVID-19 recovered individuals toward wild-type and N121Q mutated spike protein. The findings revealed that about 17% of IgGs fail to bind with wild-type spike protein in presence of biliverdin. Interestingly, biliverdin could not affect the binding efficiency of antibodies developed specifically against the spike RBD. Moreover, it was observed that biliverdin significantly reduces the neutralization of SARS-CoV-2 by a panel of neutralizing antibodies.
Regarding the mode of action of biliverdin, the scientists noticed that by binding to the S1 pocket, biliverdin restricts the relocation of the gating loop required for antibody binding to its epitope. In other words, biliverdin prevents the spike- antibody binding by restricting the conformational remodeling of the spike N-terminal domain.
Study significance
The study reveals that biliverdin, a heme metabolism product, can facilitate SARS-CoV-2 to escape antibody-mediated host immune responses. Moreover, the study identifies a neutralizing epitope on the spike N-terminal domain that is differentially exposed to neutralizing antibodies based on the recruitment of biliverdin.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources